Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A Protein Interaction Information-based Generative Model for Enhancing Gene Clustering.
Scientific Reports ( IF 3.8 ) Pub Date : 2020-01-20 , DOI: 10.1038/s41598-020-57437-5 Pratik Dutta 1 , Sriparna Saha 1 , Sanket Pai 2 , Aviral Kumar 2
Scientific Reports ( IF 3.8 ) Pub Date : 2020-01-20 , DOI: 10.1038/s41598-020-57437-5 Pratik Dutta 1 , Sriparna Saha 1 , Sanket Pai 2 , Aviral Kumar 2
Affiliation
|
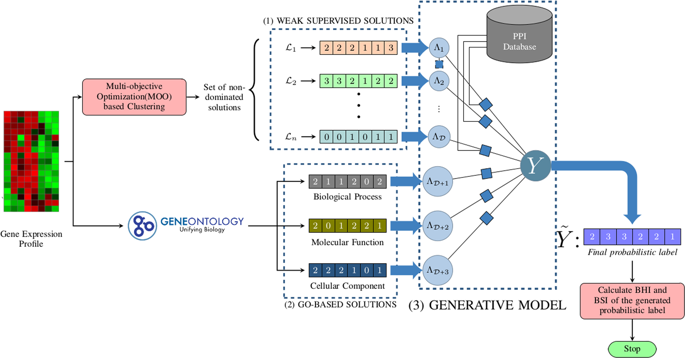
In the field of computational bioinformatics, identifying a set of genes which are responsible for a particular cellular mechanism, is very much essential for tasks such as medical diagnosis or disease gene identification. Accurately grouping (clustering) the genes is one of the important tasks in understanding the functionalities of the disease genes. In this regard, ensemble clustering becomes a promising approach to combine different clustering solutions to generate almost accurate gene partitioning. Recently, researchers have used generative model as a smart ensemble method to produce the right consensus solution. In the current paper, we develop a protein-protein interaction-based generative model that can efficiently perform a gene clustering. Utilizing protein interaction information as the generative model's latent variable enables enhance the generative model's efficiency in inferring final probabilistic labels. The proposed generative model utilizes different weak supervision sources rather utilizing any ground truth information. For weak supervision sources, we use a multi-objective optimization based clustering technique together with the world's largest gene ontology based knowledge-base named Gene Ontology Consortium(GOC). These weakly supervised labels are supplied to a generative model that eventually assigns all genes to probabilistic labels. The comparative study with respect to silhouette score, Biological Homogeneity Index (BHI) and Biological Stability Index (BSI) proves that the proposed generative model outperforms than other state-of-the-art techniques.
中文翻译:

用于增强基因聚类的基于蛋白质相互作用信息的生成模型。
在计算生物信息学领域,识别一组负责特定细胞机制的基因对于医学诊断或疾病基因识别等任务非常重要。准确地对基因进行分组(聚类)是了解疾病基因功能的重要任务之一。在这方面,集成聚类成为一种有前途的方法,可以结合不同的聚类解决方案来生成几乎准确的基因分区。最近,研究人员使用生成模型作为智能集成方法来产生正确的共识解决方案。在本文中,我们开发了一种基于蛋白质-蛋白质相互作用的生成模型,可以有效地执行基因聚类。利用蛋白质相互作用信息作为生成模型的潜在变量可以提高生成模型推断最终概率标签的效率。所提出的生成模型利用不同的弱监督源,而不是利用任何地面实况信息。对于弱监督源,我们使用基于多目标优化的聚类技术以及世界上最大的基于基因本体的知识库——基因本体联盟(GOC)。这些弱监督标签被提供给生成模型,该模型最终将所有基因分配给概率标签。轮廓评分、生物均匀性指数(BHI)和生物稳定性指数(BSI)的比较研究证明,所提出的生成模型优于其他最先进的技术。
更新日期:2020-01-21
中文翻译:

用于增强基因聚类的基于蛋白质相互作用信息的生成模型。
在计算生物信息学领域,识别一组负责特定细胞机制的基因对于医学诊断或疾病基因识别等任务非常重要。准确地对基因进行分组(聚类)是了解疾病基因功能的重要任务之一。在这方面,集成聚类成为一种有前途的方法,可以结合不同的聚类解决方案来生成几乎准确的基因分区。最近,研究人员使用生成模型作为智能集成方法来产生正确的共识解决方案。在本文中,我们开发了一种基于蛋白质-蛋白质相互作用的生成模型,可以有效地执行基因聚类。利用蛋白质相互作用信息作为生成模型的潜在变量可以提高生成模型推断最终概率标签的效率。所提出的生成模型利用不同的弱监督源,而不是利用任何地面实况信息。对于弱监督源,我们使用基于多目标优化的聚类技术以及世界上最大的基于基因本体的知识库——基因本体联盟(GOC)。这些弱监督标签被提供给生成模型,该模型最终将所有基因分配给概率标签。轮廓评分、生物均匀性指数(BHI)和生物稳定性指数(BSI)的比较研究证明,所提出的生成模型优于其他最先进的技术。











































 京公网安备 11010802027423号
京公网安备 11010802027423号