当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Why Purine Nucleoside Phosphorylase Ribosylates 2,6-Diamino-8-azapurine in Noncanonical Positions? A Molecular Modeling Study.
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-01-16 , DOI: 10.1021/acs.jcim.9b00985 Maciej Pyrka 1 , Maciej Maciejczyk 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2020-01-16 , DOI: 10.1021/acs.jcim.9b00985 Maciej Pyrka 1 , Maciej Maciejczyk 1
Affiliation
|
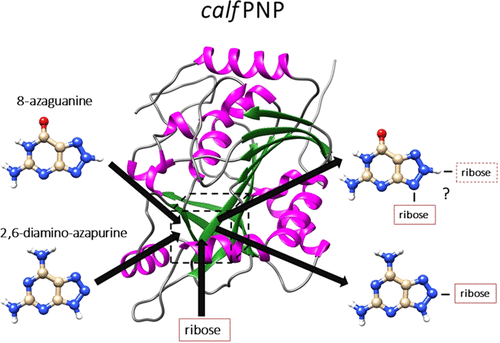
Protein nucleoside phosphorylase (PNP) is an enzyme that catalyzes a reversible conversion process (ribosylation and phosphorolysis) between nucleobases (purines) and their nucleosides. Experimental studies showed that calf PNP ribosylates purine analogues in specific positions: 2,6-diamino-8-azapurine in position 7 or 8 and 8-azaguanine in position 9 of the triazole ring. The reason for this phenomenon can be a result of different expositions of purine substrates to the channel leading to the binding site. This hypothesis was verified by the application of molecular modeling techniques to two complexes of purine analogues 2,6-diamino-azapurine, calf PNP (pdb-code: 1LVU ), and 8-azaguanine, calf PNP (pdb-code: 2AI1). The results obtained with a combination of quantum chemistry, docking, and molecular dynamics methods showed qualitative validity of our hypothesis. Binding free energies of protein-ligand systems showed that most probable binding poses expose N8 nitrogen for 2,6-diamino-8-azapurine and N9 nitrogen for 8-azaguanine into the binding channel and ruled out the exposition of N9 for 2,6-diamino-8-azapurine and N7 for 8-azaguanine, partially in agreement with the experimental data. The other important result obtained in this study is a significantly higher population of the protonated form of crucial residue Glu-201 present in the binding pocket, compared to the standard protonation of free glutamic acid in solution. This result combined with populations of tautomeric forms of both investigated systems strongly suggests that 2,6-diamino-8-azapurine and 8-azaguanine are recognized by proteins with deprotonated and protonated Glu-201 residues, respectively. A comparison of computed binding poses of the investigated ligands to the inhibitors present in crystal structures suggests that the modification of the (S)-PMPDAP inhibitor, in which a 2-(phosphonomethoxy)propyl chain is attached at position 8 instead of position 9, might increase its binding affinity.
中文翻译:

为什么嘌呤核苷磷酸化酶核糖基化非常规位置的2,6-二氨基-8-氮杂嘌呤?分子建模研究。
蛋白质核苷磷酸化酶(PNP)是一种催化核碱基(嘌呤)与其核苷之间可逆转化过程(核糖基化和磷酸分解)的酶。实验研究表明,小牛PNP会在特定位置将嘌呤类似物核糖基化:在三唑环的第7或8位为2,6-二氨基-8-氮杂嘌呤,在第9位置为8-氮杂鸟嘌呤。产生这种现象的原因可能是由于嘌呤底物暴露于导致结合位点的通道的方式不同。通过将分子建模技术应用于嘌呤类似物2,6-二氨基-氮杂嘌呤,小牛PNP(pdb-code:1LVU)和8-氮杂鸟嘌呤,小牛PNP(pdb-code:2AI1)的两种复合物,证实了这一假设。结合量子化学,对接,分子动力学方法证明了我们假设的定性有效性。蛋白质-配体系统的结合自由能表明,大多数可能的结合姿势将2,6-二氨基-8-氮杂嘌呤的N8氮和8-氮杂鸟嘌呤的N9氮暴露于结合通道中,并排除了N9暴露于2,6-二氨基8-氮杂嘌呤和N7代表8-氮杂鸟嘌呤,部分与实验数据一致。在这项研究中获得的另一个重要结果是,与溶液中游离谷氨酸的标准质子化相比,结合口袋中存在的重要残基Glu-201的质子化形式的数量明显增加。该结果与两个系统的互变异构形式的总体结合强烈表明,2 6-二氨基-8-氮杂嘌呤和8-氮杂鸟嘌呤分别被具有去质子化和质子化的Glu-201残基的蛋白质识别。比较所研究的配体与晶体结构中存在的抑制剂的结合姿势,表明(S)-PMPDAP抑制剂的修饰形式,其中2-(膦酰基甲氧基)丙基链连接在位置8而不是位置9,可能会增加其结合亲和力。
更新日期:2020-01-16
中文翻译:

为什么嘌呤核苷磷酸化酶核糖基化非常规位置的2,6-二氨基-8-氮杂嘌呤?分子建模研究。
蛋白质核苷磷酸化酶(PNP)是一种催化核碱基(嘌呤)与其核苷之间可逆转化过程(核糖基化和磷酸分解)的酶。实验研究表明,小牛PNP会在特定位置将嘌呤类似物核糖基化:在三唑环的第7或8位为2,6-二氨基-8-氮杂嘌呤,在第9位置为8-氮杂鸟嘌呤。产生这种现象的原因可能是由于嘌呤底物暴露于导致结合位点的通道的方式不同。通过将分子建模技术应用于嘌呤类似物2,6-二氨基-氮杂嘌呤,小牛PNP(pdb-code:1LVU)和8-氮杂鸟嘌呤,小牛PNP(pdb-code:2AI1)的两种复合物,证实了这一假设。结合量子化学,对接,分子动力学方法证明了我们假设的定性有效性。蛋白质-配体系统的结合自由能表明,大多数可能的结合姿势将2,6-二氨基-8-氮杂嘌呤的N8氮和8-氮杂鸟嘌呤的N9氮暴露于结合通道中,并排除了N9暴露于2,6-二氨基8-氮杂嘌呤和N7代表8-氮杂鸟嘌呤,部分与实验数据一致。在这项研究中获得的另一个重要结果是,与溶液中游离谷氨酸的标准质子化相比,结合口袋中存在的重要残基Glu-201的质子化形式的数量明显增加。该结果与两个系统的互变异构形式的总体结合强烈表明,2 6-二氨基-8-氮杂嘌呤和8-氮杂鸟嘌呤分别被具有去质子化和质子化的Glu-201残基的蛋白质识别。比较所研究的配体与晶体结构中存在的抑制剂的结合姿势,表明(S)-PMPDAP抑制剂的修饰形式,其中2-(膦酰基甲氧基)丙基链连接在位置8而不是位置9,可能会增加其结合亲和力。











































 京公网安备 11010802027423号
京公网安备 11010802027423号