iScience ( IF 4.6 ) Pub Date : 2020-01-16 , DOI: 10.1016/j.isci.2020.100842 Denis Dermadi 1 , Michael Bscheider 1 , Kristina Bjegovic 2 , Nicole H Lazarus 1 , Agata Szade 1 , Husein Hadeiba 2 , Eugene C Butcher 1
|
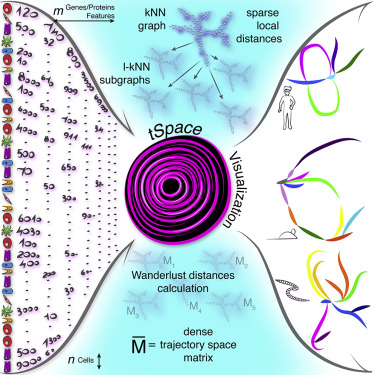
High-dimensional single cell profiling coupled with computational modeling is emerging as a powerful tool to elucidate developmental programs directing cell lineages. We introduce tSpace, an algorithm based on the concept of “trajectory space”, in which cells are defined by their distance along nearest neighbor pathways to every other cell in a population. Graphical mapping of cells in trajectory space allows unsupervised reconstruction and exploration of complex developmental sequences. Applied to flow and mass cytometry data, the method faithfully reconstructs thymic T cell development and reveals development and trafficking regulation of tonsillar B cells. Applied to the single cell transcriptome of mouse intestine and C. elegans, the method recapitulates development from intestinal stem cells to specialized epithelial phenotypes more faithfully than existing algorithms and orders C. elegans cells concordantly to the associated embryonic time. tSpace profiling of complex populations is well suited for hypothesis generation in developing cell systems.
中文翻译:

通过轨迹空间中的高维单细胞分析探索细胞发育途径。
高维单细胞分析与计算建模相结合正在成为阐明指导细胞谱系的发育程序的强大工具。我们引入了 tSpace,这是一种基于“轨迹空间”概念的算法,其中细胞是根据它们沿着最近邻路径到群体中每个其他细胞的距离来定义的。轨迹空间中细胞的图形映射允许无监督地重建和探索复杂的发育序列。该方法应用于流式细胞术和质谱流式细胞术数据,忠实地重建了胸腺 T 细胞的发育,并揭示了扁桃体 B 细胞的发育和运输调控。该方法应用于小鼠肠道和线虫的单细胞转录组,比现有算法更忠实地概括了从肠道干细胞到特殊上皮表型的发育,并对线虫细胞进行了与相关胚胎时间一致的排序。复杂群体的 tSpace 分析非常适合在发育中的细胞系统中生成假设。











































 京公网安备 11010802027423号
京公网安备 11010802027423号