当前位置:
X-MOL 学术
›
Nat. Commun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Intratumoral heterogeneity and clonal evolution in liver cancer.
Nature Communications ( IF 14.7 ) Pub Date : 2020-01-15 , DOI: 10.1038/s41467-019-14050-z Bojan Losic 1, 2, 3 , Amanda J Craig 4 , Carlos Villacorta-Martin 4 , Sebastiao N Martins-Filho 4, 5 , Nicholas Akers 1, 6 , Xintong Chen 1 , Mehmet E Ahsen 1 , Johann von Felden 4, 7 , Ismail Labgaa 4, 8 , Delia DʹAvola 4, 9 , Kimaada Allette 1, 2 , Sergio A Lira 10 , Glaucia C Furtado 10 , Teresa Garcia-Lezana 4 , Paula Restrepo 1 , Ashley Stueck 11 , Stephen C Ward 12 , Maria I Fiel 12 , Spiros P Hiotis 13 , Ganesh Gunasekaran 13 , Daniela Sia 4 , Eric E Schadt 2, 14 , Robert Sebra 1, 2, 14 , Myron Schwartz 13 , Josep M Llovet 4, 15, 16 , Swan Thung 12 , Gustavo Stolovitzky 1, 17 , Augusto Villanueva 4, 18
Nature Communications ( IF 14.7 ) Pub Date : 2020-01-15 , DOI: 10.1038/s41467-019-14050-z Bojan Losic 1, 2, 3 , Amanda J Craig 4 , Carlos Villacorta-Martin 4 , Sebastiao N Martins-Filho 4, 5 , Nicholas Akers 1, 6 , Xintong Chen 1 , Mehmet E Ahsen 1 , Johann von Felden 4, 7 , Ismail Labgaa 4, 8 , Delia DʹAvola 4, 9 , Kimaada Allette 1, 2 , Sergio A Lira 10 , Glaucia C Furtado 10 , Teresa Garcia-Lezana 4 , Paula Restrepo 1 , Ashley Stueck 11 , Stephen C Ward 12 , Maria I Fiel 12 , Spiros P Hiotis 13 , Ganesh Gunasekaran 13 , Daniela Sia 4 , Eric E Schadt 2, 14 , Robert Sebra 1, 2, 14 , Myron Schwartz 13 , Josep M Llovet 4, 15, 16 , Swan Thung 12 , Gustavo Stolovitzky 1, 17 , Augusto Villanueva 4, 18
Affiliation
|
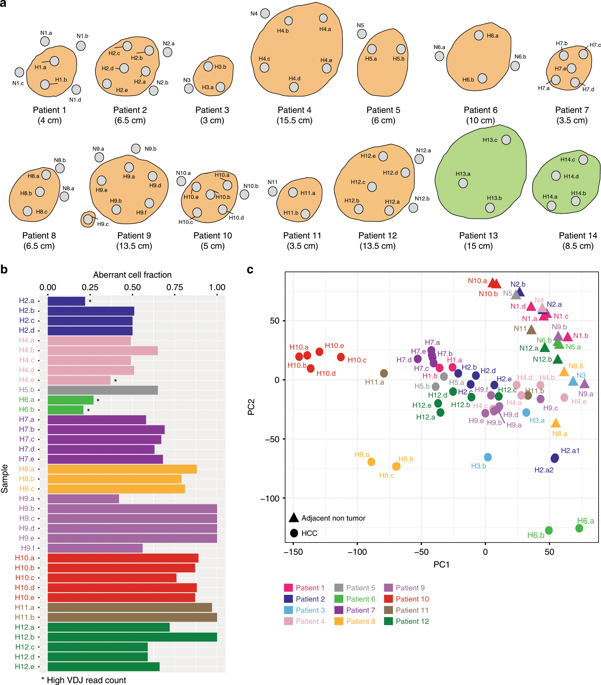
Clonal evolution of a tumor ecosystem depends on different selection pressures that are principally immune and treatment mediated. We integrate RNA-seq, DNA sequencing, TCR-seq and SNP array data across multiple regions of liver cancer specimens to map spatio-temporal interactions between cancer and immune cells. We investigate how these interactions reflect intra-tumor heterogeneity (ITH) by correlating regional neo-epitope and viral antigen burden with the regional adaptive immune response. Regional expression of passenger mutations dominantly recruits adaptive responses as opposed to hepatitis B virus and cancer-testis antigens. We detect different clonal expansion of the adaptive immune system in distant regions of the same tumor. An ITH-based gene signature improves single-biopsy patient survival predictions and an expression survey of 38,553 single cells across 7 regions of 2 patients further reveals heterogeneity in liver cancer. These data quantify transcriptomic ITH and how the different components of the HCC ecosystem interact during cancer evolution.
中文翻译:

肝癌的瘤内异质性和克隆进化。
肿瘤生态系统的克隆进化取决于主要是免疫和治疗介导的不同选择压力。我们整合了肝癌样本多个区域的 RNA-seq、DNA 测序、TCR-seq 和 SNP 阵列数据,以绘制癌症和免疫细胞之间的时空相互作用。我们通过将区域新表位和病毒抗原负荷与区域适应性免疫反应相关联,研究这些相互作用如何反映肿瘤内异质性 (ITH)。与乙型肝炎病毒和癌症睾丸抗原相反,乘客突变的区域表达主要招募适应性反应。我们在同一肿瘤的远处区域检测到适应性免疫系统的不同克隆扩增。基于 ITH 的基因特征提高了单次活检患者的生存预测和 38 人的表达调查,2 名患者 7 个区域的 553 个单细胞进一步揭示了肝癌的异质性。这些数据量化了转录组 ITH 以及 HCC 生态系统的不同组成部分在癌症进化过程中如何相互作用。
更新日期:2020-01-15
中文翻译:

肝癌的瘤内异质性和克隆进化。
肿瘤生态系统的克隆进化取决于主要是免疫和治疗介导的不同选择压力。我们整合了肝癌样本多个区域的 RNA-seq、DNA 测序、TCR-seq 和 SNP 阵列数据,以绘制癌症和免疫细胞之间的时空相互作用。我们通过将区域新表位和病毒抗原负荷与区域适应性免疫反应相关联,研究这些相互作用如何反映肿瘤内异质性 (ITH)。与乙型肝炎病毒和癌症睾丸抗原相反,乘客突变的区域表达主要招募适应性反应。我们在同一肿瘤的远处区域检测到适应性免疫系统的不同克隆扩增。基于 ITH 的基因特征提高了单次活检患者的生存预测和 38 人的表达调查,2 名患者 7 个区域的 553 个单细胞进一步揭示了肝癌的异质性。这些数据量化了转录组 ITH 以及 HCC 生态系统的不同组成部分在癌症进化过程中如何相互作用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号