Nature Biotechnology ( IF 33.1 ) Pub Date : 2020-01-13 , DOI: 10.1038/s41587-019-0393-7 Chao Li 1, 2 , Rui Zhang 1 , Xiangbing Meng 3 , Sha Chen 1, 2 , Yuan Zong 1, 2 , Chunju Lu 1, 2 , Jin-Long Qiu 2, 4 , Yu-Hang Chen 2, 5 , Jiayang Li 2, 3 , Caixia Gao 1, 2
|
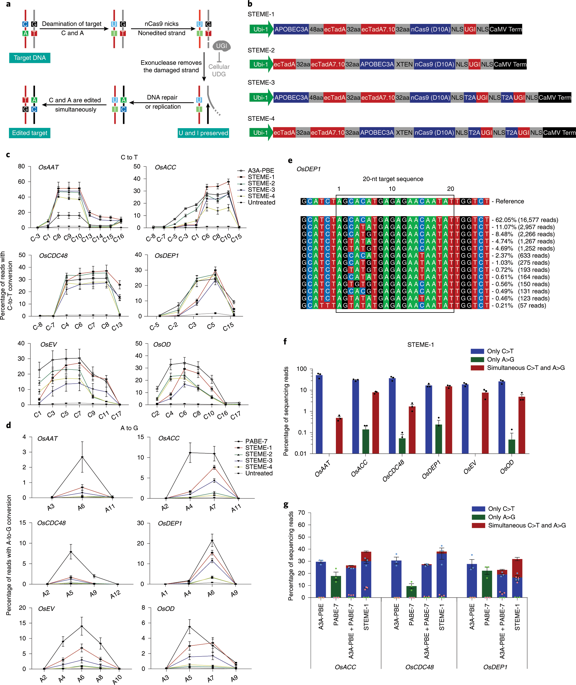
Targeted saturation mutagenesis of crop genes could be applied to produce genetic variants with improved agronomic performance. However, tools for directed evolution of plant genes, such as error-prone PCR or DNA shuffling, are limited1. We engineered five saturated targeted endogenous mutagenesis editors (STEMEs) that can generate de novo mutations and facilitate directed evolution of plant genes. In rice protoplasts, STEME-1 edited cytosine and adenine at the same target site with C > T efficiency up to 61.61% and simultaneous C > T and A > G efficiency up to 15.10%. STEME-NG, which incorporates the nickase Cas9-NG protospacer-adjacent motif variant, was used with 20 individual single guide RNAs in rice protoplasts to produce near-saturated mutagenesis (73.21%) for a 56-amino-acid portion of the rice acetyl-coenzyme A carboxylase (OsACC). We also applied STEME-1 and STEME-NG for directed evolution of the OsACC gene in rice and obtained herbicide resistance mutations. This set of two STEMEs will accelerate trait development and should work in any plants amenable to CRISPR-based editing.
中文翻译:

使用双胞嘧啶和腺嘌呤碱基编辑器对植物基因进行定向、随机诱变
作物基因的定向饱和诱变可用于产生具有改善农艺性能的遗传变体。然而,用于植物基因定向进化的工具(例如易错 PCR 或 DNA 改组)是有限的1 。我们设计了五个饱和的靶向内源诱变编辑器(STEME),它们可以产生从头突变并促进植物基因的定向进化。在水稻原生质体中,STEME-1 在同一靶位点编辑胞嘧啶和腺嘌呤,C > T 效率高达 61.61%,同时 C > T 和 A > G 效率高达 15.10%。 STEME-NG 包含切口酶 Cas9-NG 原型间隔子相邻基序变体,与水稻原生质体中的 20 个单独的单向导 RNA 一起使用,对水稻乙酰基的 56 个氨基酸部分产生近饱和诱变 (73.21%) -辅酶A羧化酶(OsACC)。我们还应用STEME-1和STEME-NG对水稻OsACC基因进行定向进化,获得了除草剂抗性突变。这组两个 STEME 将加速性状发育,并且应该适用于任何适合基于 CRISPR 编辑的植物。











































 京公网安备 11010802027423号
京公网安备 11010802027423号