Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Highly drug-resistant HIV-1 protease reveals decreased intra-subunit interactions due to clusters of mutations.
The FEBS Journal ( IF 5.5 ) Pub Date : 2020-01-09 , DOI: 10.1111/febs.15207 Daniel W Kneller 1 , Johnson Agniswamy 1 , Robert W Harrison 2 , Irene T Weber 1, 3
The FEBS Journal ( IF 5.5 ) Pub Date : 2020-01-09 , DOI: 10.1111/febs.15207 Daniel W Kneller 1 , Johnson Agniswamy 1 , Robert W Harrison 2 , Irene T Weber 1, 3
Affiliation
|
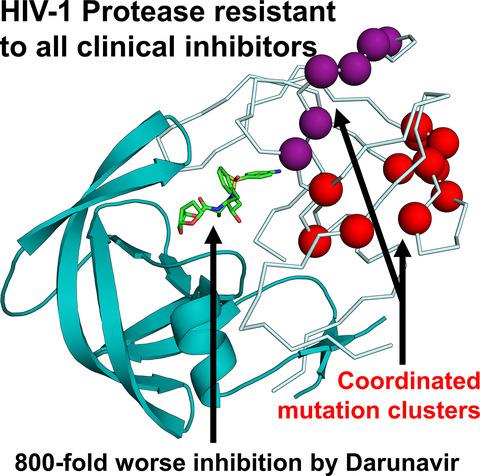
Drug‐resistance is a serious problem for treatment of the HIV/AIDS pandemic. Potent clinical inhibitors of HIV‐1 protease show several orders of magnitude worse inhibition of highly drug‐resistant variants. Hence, the structure and enzyme activities were analyzed for HIV protease mutant HIV‐1 protease (EC 3.4.23.16) (PR) with 22 mutations (PRS5B) from a clinical isolate that was selected by machine learning to represent high‐level drug‐resistance. PRS5B has 22 mutations including only one (I84V) in the inhibitor binding site; however, clinical inhibitors had poor inhibition of PRS5B activity with kinetic inhibition value (Ki ) values of 4–1000 nm or 18‐ to 8000‐fold worse than for wild‐type PR. High‐resolution crystal structures of PRS5B complexes with the best inhibitors, amprenavir (APV) and darunavir (DRV) (Ki ~ 4 nm ), revealed only minor changes in protease‐inhibitor interactions. Instead, two distinct clusters of mutations in distal regions induce coordinated conformational changes that decrease favorable internal interactions across the entire protein subunit. The largest structural rearrangements are described and compared to other characterized resistant mutants. In the protease hinge region, the N83D mutation eliminates a hydrogen bond connecting the hinge and core of the protease and increases disorder compared to highly resistant mutants PR with 17 mutations and PR with 20 mutations with similar hinge mutations. In a distal β‐sheet, mutations G73T and A71V coordinate with accessory mutations to bring about shifts that propagate throughout the subunit. Molecular dynamics simulations of ligand‐free dimers show differences consistent with loss of interactions in mutant compared to wild‐type PR. Clusters of mutations exhibit both coordinated and antagonistic effects, suggesting PRS5B may represent an intermediate stage in the evolution of more highly resistant variants.
中文翻译:

高度耐药的 HIV-1 蛋白酶显示由于突变簇导致亚基内相互作用减少。
耐药性是治疗 HIV/AIDS 大流行病的一个严重问题。HIV-1 蛋白酶的有效临床抑制剂对高度耐药变体的抑制效果差几个数量级。因此,对具有 22 个突变 (PRS5B) 的 HIV 蛋白酶突变体 HIV-1 蛋白酶 (EC 3.4.23.16) (PR) 的结构和酶活性进行了分析,该突变体来自通过机器学习选择的代表高水平耐药性的临床分离株. PRS5B 有 22 个突变,其中只有一个 (I84V) 在抑制剂结合位点;然而,临床抑制剂对 PRS5B 活性的抑制作用较差,动力学抑制值 ( K i ) 值为 4–1000 nm或比野生型 PR 差 18 到 8000 倍。PRS5B 复合物与最佳抑制剂安普那韦 (APV) 和地瑞那韦 (DRV) 的高分辨率晶体结构 ( K i ~ 4 n m), 仅显示蛋白酶-抑制剂相互作用的微小变化。相反,远端区域的两个不同的突变簇会引起协调的构象变化,从而减少整个蛋白质亚基之间有利的内部相互作用。描述了最大的结构重排并与其他表征的抗性突变体进行了比较。在蛋白酶铰链区,N83D 突变消除了连接蛋白酶铰链和核心的氢键,与具有 17 个突变的高耐药突变体 PR 和具有类似铰链突变的 20 个突变的 PR 相比,增加了无序。在远端 β 片层中,突变 G73T 和 A71V 与附属突变相协调,导致在整个亚基中传播的变化。与野生型 PR 相比,无配体二聚体的分子动力学模拟显示与突变体相互作用丧失一致的差异。突变簇表现出协调和拮抗作用,表明 PRS5B 可能代表更高抗性变体进化的中间阶段。
更新日期:2020-01-09
中文翻译:

高度耐药的 HIV-1 蛋白酶显示由于突变簇导致亚基内相互作用减少。
耐药性是治疗 HIV/AIDS 大流行病的一个严重问题。HIV-1 蛋白酶的有效临床抑制剂对高度耐药变体的抑制效果差几个数量级。因此,对具有 22 个突变 (PRS5B) 的 HIV 蛋白酶突变体 HIV-1 蛋白酶 (EC 3.4.23.16) (PR) 的结构和酶活性进行了分析,该突变体来自通过机器学习选择的代表高水平耐药性的临床分离株. PRS5B 有 22 个突变,其中只有一个 (I84V) 在抑制剂结合位点;然而,临床抑制剂对 PRS5B 活性的抑制作用较差,动力学抑制值 ( K i ) 值为 4–1000 nm或比野生型 PR 差 18 到 8000 倍。PRS5B 复合物与最佳抑制剂安普那韦 (APV) 和地瑞那韦 (DRV) 的高分辨率晶体结构 ( K i ~ 4 n m), 仅显示蛋白酶-抑制剂相互作用的微小变化。相反,远端区域的两个不同的突变簇会引起协调的构象变化,从而减少整个蛋白质亚基之间有利的内部相互作用。描述了最大的结构重排并与其他表征的抗性突变体进行了比较。在蛋白酶铰链区,N83D 突变消除了连接蛋白酶铰链和核心的氢键,与具有 17 个突变的高耐药突变体 PR 和具有类似铰链突变的 20 个突变的 PR 相比,增加了无序。在远端 β 片层中,突变 G73T 和 A71V 与附属突变相协调,导致在整个亚基中传播的变化。与野生型 PR 相比,无配体二聚体的分子动力学模拟显示与突变体相互作用丧失一致的差异。突变簇表现出协调和拮抗作用,表明 PRS5B 可能代表更高抗性变体进化的中间阶段。











































 京公网安备 11010802027423号
京公网安备 11010802027423号