Nature Biotechnology ( IF 33.1 ) Pub Date : 2020-01-09 , DOI: 10.1038/s41587-019-0364-z Adriana Salcedo 1, 2 , Maxime Tarabichi 3, 4 , Shadrielle Melijah G Espiritu 1 , Amit G Deshwar 5 , Matei David 1 , Nathan M Wilson 1 , Stefan Dentro 3, 4 , Jeff A Wintersinger 6 , Lydia Y Liu 1 , Minjeong Ko 1 , Srinivasan Sivanandan 1 , Hongjiu Zhang 7 , Kaiyi Zhu 8, 9, 10 , Tai-Hsien Ou Yang 8, 9, 10 , John M Chilton 11 , Alex Buchanan 12 , Christopher M Lalansingh 1 , Christine P'ng 1 , Catalina V Anghel 1 , Imaad Umar 1 , Bryan Lo 1 , William Zou 1 , , Jared T Simpson 1 , Joshua M Stuart 13 , Dimitris Anastassiou 8, 9, 10, 14 , Yuanfang Guan 7, 15, 16 , Adam D Ewing 17 , Kyle Ellrott 11, 12 , David C Wedge 18, 19 , Quaid Morris 1, 6, 20, 21 , Peter Van Loo 3, 22 , Paul C Boutros 2, 23, 24, 25, 26, 27
|
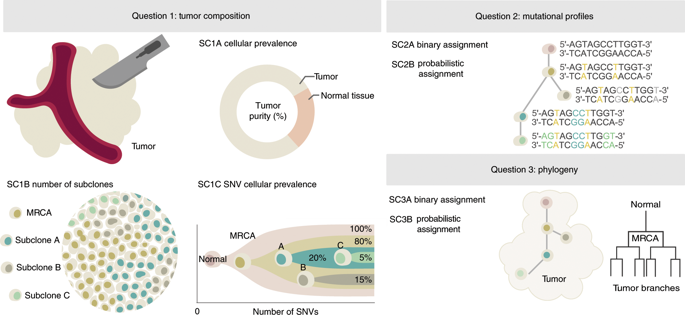
Tumor DNA sequencing data can be interpreted by computational methods that analyze genomic heterogeneity to infer evolutionary dynamics. A growing number of studies have used these approaches to link cancer evolution with clinical progression and response to therapy. Although the inference of tumor phylogenies is rapidly becoming standard practice in cancer genome analyses, standards for evaluating them are lacking. To address this need, we systematically assess methods for reconstructing tumor subclonality. First, we elucidate the main algorithmic problems in subclonal reconstruction and develop quantitative metrics for evaluating them. Then we simulate realistic tumor genomes that harbor all known clonal and subclonal mutation types and processes. Finally, we benchmark 580 tumor reconstructions, varying tumor read depth, tumor type and somatic variant detection. Our analysis provides a baseline for the establishment of gold-standard methods to analyze tumor heterogeneity.
中文翻译:

社区努力制定评估肿瘤亚克隆重建的标准
肿瘤 DNA 测序数据可以通过分析基因组异质性的计算方法来解释,从而推断进化动态。越来越多的研究使用这些方法将癌症演变与临床进展和治疗反应联系起来。尽管肿瘤系统发育的推断正在迅速成为癌症基因组分析的标准做法,但缺乏评估它们的标准。为了满足这一需求,我们系统地评估了重建肿瘤亚克隆性的方法。首先,我们阐明了亚克隆重建中的主要算法问题,并制定了评估它们的定量指标。然后,我们模拟真实的肿瘤基因组,其中包含所有已知的克隆和亚克隆突变类型和过程。最后,我们对 580 个肿瘤重建、不同的肿瘤读取深度、肿瘤类型和体细胞变异检测进行了基准测试。我们的分析为建立分析肿瘤异质性的金标准方法提供了基线。










































 京公网安备 11010802027423号
京公网安备 11010802027423号