iScience ( IF 4.6 ) Pub Date : 2020-01-10 , DOI: 10.1016/j.isci.2020.100824 Max Schnepf 1 , Claudia Ludwig 1 , Peter Bandilla 1 , Stefano Ceolin 1 , Ulrich Unnerstall 1 , Christophe Jung 1 , Ulrike Gaul 1
|
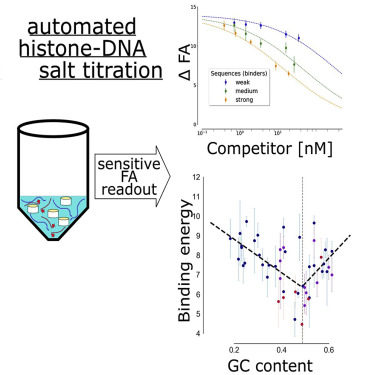
The DNA of eukaryotes is wrapped around histone octamers to form nucleosomes. Although it is well established that the DNA sequence significantly influences nucleosome formation, its precise contribution has remained controversial, partially owing to the lack of quantitative affinity data. Here, we present a method to measure DNA-histone binding free energies at medium throughput and with high sensitivity. Competitive nucleosome formation is achieved through automation, and a modified epifluorescence microscope is used to rapidly and accurately measure the fractions of bound/unbound DNA based on fluorescence anisotropy. The procedure allows us to obtain full titration curves with high reproducibility. We applied this technique to measure the histone-DNA affinities for 47 DNA sequences and analyzed how the affinities correlate with relevant DNA sequence features. We found that the GC content has a significant impact on nucleosome-forming preferences, but 10 bp dinucleotide periodicities and the presence of poly(dA:dT) stretches do not.
中文翻译:

核糖体中组蛋白-DNA亲和力的灵敏自动测量。
真核生物的DNA包裹在组蛋白八聚体周围,形成核小体。尽管众所周知,DNA序列会显着影响核小体的形成,但其精确的贡献仍存在争议,部分原因是缺乏定量亲和力数据。在这里,我们提出了一种以中等吞吐量和高灵敏度测量DNA-组蛋白结合自由能的方法。竞争性核小体的形成是通过自动化实现的,基于荧光各向异性,使用改良的落射荧光显微镜可以快速,准确地测量结合/未结合的DNA组分。该程序使我们可以获得具有高重现性的完整滴定曲线。我们应用了该技术来测量47个DNA序列的组蛋白-DNA亲和力,并分析了亲和力如何与相关的DNA序列特征相关。我们发现,GC含量对形成核小体的偏好有重大影响,但10 bp的二核苷酸周期和poly(dA:dT)延伸的存在却没有。










































 京公网安备 11010802027423号
京公网安备 11010802027423号