当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Designed Alteration of Binding Affinity in Structure-Switching Aptamers through the Use of Dangling Nucleotides.
Biochemistry ( IF 2.9 ) Pub Date : 2020-01-16 , DOI: 10.1021/acs.biochem.9b00630 Sladjana Slavkovic 1 , Sophie R Eisen 1 , Philip E Johnson 1
Biochemistry ( IF 2.9 ) Pub Date : 2020-01-16 , DOI: 10.1021/acs.biochem.9b00630 Sladjana Slavkovic 1 , Sophie R Eisen 1 , Philip E Johnson 1
Affiliation
|
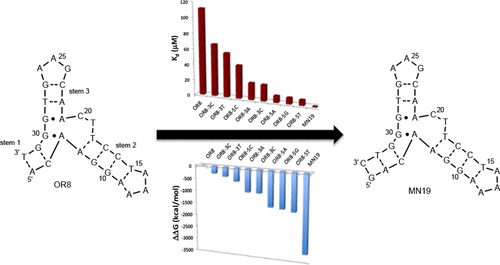
The ability to change binding affinity in a controlled fashion is a key step in the rational design of biomolecules in general and functional nucleic acids in particular. Here, we use dangling nucleotides to alter the binding affinity of structure-switching aptamers. Dangling nucleotides can stabilize or destabilize a nucleic acid structure with a known ΔG°37. When the dangling nucleotide stabilizes the structure, less free energy from ligand binding is needed to fold the molecule and hence the ligand is observed to bind tighter than in the absence of the unpaired nucleotide. For a destabilizing dangling nucleotide, the opposite occurs, and the observed binding is weaker. We demonstrate this concept using both the cocaine-binding aptamer and the ATP-binding aptamer systems. We find that for both aptamers there is a direct, but different, relationship between the predicted stabilization and the change in the observed binding free energy.
中文翻译:

通过使用悬挂的核苷酸设计改变结构转换适体中的结合亲和力。
以受控方式改变结合亲和力的能力是合理设计生物分子的关键步骤,尤其是功能核酸。在这里,我们使用悬空的核苷酸来改变结构转换适体的结合亲和力。悬空的核苷酸可以用已知的ΔG°37稳定或破坏核酸结构。当悬空的核苷酸使结构稳定时,需要较少的来自配体结合的自由能来折叠分子,因此观察到的配体与不存在未配对核苷酸的情况下结合更紧密。对于不稳定的悬空核苷酸,则相反,观察到的结合较弱。我们使用可卡因结合的适体和ATP结合的适体系统证明了这一概念。我们发现,对于两种适体,都有直接但不同的
更新日期:2020-01-17
中文翻译:

通过使用悬挂的核苷酸设计改变结构转换适体中的结合亲和力。
以受控方式改变结合亲和力的能力是合理设计生物分子的关键步骤,尤其是功能核酸。在这里,我们使用悬空的核苷酸来改变结构转换适体的结合亲和力。悬空的核苷酸可以用已知的ΔG°37稳定或破坏核酸结构。当悬空的核苷酸使结构稳定时,需要较少的来自配体结合的自由能来折叠分子,因此观察到的配体与不存在未配对核苷酸的情况下结合更紧密。对于不稳定的悬空核苷酸,则相反,观察到的结合较弱。我们使用可卡因结合的适体和ATP结合的适体系统证明了这一概念。我们发现,对于两种适体,都有直接但不同的











































 京公网安备 11010802027423号
京公网安备 11010802027423号