当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Calibration of the dianionic phosphate group: Validation on the recognition site of the homodimeric enzyme phosphoglucose isomerase
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2020-01-07 , DOI: 10.1002/jcc.26134 Marion Devillers 1 , Jean-Philip Piquemal 2, 3 , Laurent Salmon 1 , Nohad Gresh 2
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2020-01-07 , DOI: 10.1002/jcc.26134 Marion Devillers 1 , Jean-Philip Piquemal 2, 3 , Laurent Salmon 1 , Nohad Gresh 2
Affiliation
|
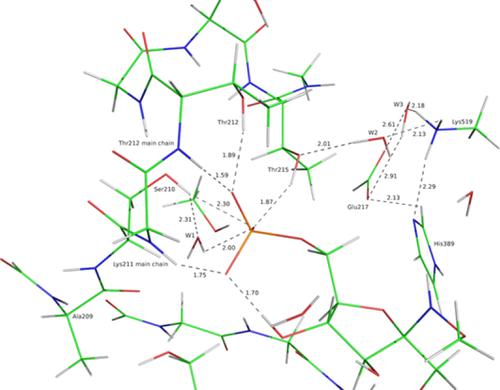
We calibrate and validate the parameters necessary to represent the dianionic phosphate group (DPG) in molecular mechanics. DPG is an essential fragment of signaling biological molecules and protein‐binding ligands. It is a constitutive fragment of biosensors, which bind to the dimer interface of phosphoglucose isomerase (PGI), an intracellular enzyme involved in sugar metabolism, as well as an extracellular protein known as autocrine motility factor (AMF) closely related to metastasis formation. Our long‐term objective is to design DPG‐based biosensors with enhanced affinities for AMF/PGI cancer biomarker in blood. Molecular dynamics with polarizable potentials could be used toward this aim. This requires to first evaluate the accuracy of such potentials upon representing the interactions of DPG with its PGI ligands and tightly bound water molecules. Such evaluations are done by comparisons with high‐level ab initio quantum chemistry (QC) calculations. We focus on the Sum of Interactions Between Fragments Ab initio computed (SIBFA) polarizable molecular mechanics procedure. We present first the results of the DPG calibration. This is followed by comparisons between ΔE(SIBFA) and ΔE(QC) regarding bi‐molecular complexes of DPG with the main‐chain and side‐chain PGI residues, which bind to it in the recognition site. We then consider DPG complexes with an increasing number of PGI residues. The largest QC complexes encompass the entirety of the recognition site, with six structural water molecules totaling up to 211 atoms. A persistent and satisfactory agreement could be shown between ΔE(SIBFA) and ΔE(QC). These validations constitute an essential first step toward large‐scale molecular dynamics simulations of DPG‐based biosensors bound at the PGI dimer interface. © 2020 Wiley Periodicals, Inc.
中文翻译:

双阴离子磷酸基团的校准:对同型二聚酶磷酸葡萄糖异构酶的识别位点的验证
我们校准和验证了在分子力学中表示双阴离子磷酸基团 (DPG) 所需的参数。DPG 是信号生物分子和蛋白质结合配体的重要片段。它是生物传感器的组成性片段,它与磷酸葡萄糖异构酶 (PGI) 的二聚体界面结合,PGI 是一种参与糖代谢的细胞内酶,以及与转移形成密切相关的称为自分泌运动因子 (AMF) 的细胞外蛋白。我们的长期目标是设计对血液中 AMF/PGI 癌症生物标志物具有增强亲和力的基于 DPG 的生物传感器。具有可极化电位的分子动力学可用于实现这一目标。这需要首先评估这种电位在表示 DPG 与其 PGI 配体和紧密结合的水分子之间的相互作用时的准确性。此类评估是通过与高级从头算量子化学 (QC) 计算进行比较来完成的。我们专注于片段从头计算 (SIBFA) 极化分子力学程序之间的相互作用总和。我们首先介绍 DPG 校准的结果。随后是关于 DPG 与主链和侧链 PGI 残基的双分子复合物的 ΔE(SIBFA) 和 ΔE(QC) 之间的比较,后者在识别位点与其结合。然后我们考虑具有越来越多的 PGI 残基的 DPG 复合物。最大的 QC 复合物包含整个识别位点,六个结构水分子总共多达 211 个原子。ΔE(SIBFA) 和 ΔE(QC) 之间可以显示出持久且令人满意的一致性。这些验证构成了对结合在 PGI 二聚体界面上的基于 DPG 的生物传感器进行大规模分子动力学模拟的重要第一步。© 2020 威利期刊公司。
更新日期:2020-01-07
中文翻译:

双阴离子磷酸基团的校准:对同型二聚酶磷酸葡萄糖异构酶的识别位点的验证
我们校准和验证了在分子力学中表示双阴离子磷酸基团 (DPG) 所需的参数。DPG 是信号生物分子和蛋白质结合配体的重要片段。它是生物传感器的组成性片段,它与磷酸葡萄糖异构酶 (PGI) 的二聚体界面结合,PGI 是一种参与糖代谢的细胞内酶,以及与转移形成密切相关的称为自分泌运动因子 (AMF) 的细胞外蛋白。我们的长期目标是设计对血液中 AMF/PGI 癌症生物标志物具有增强亲和力的基于 DPG 的生物传感器。具有可极化电位的分子动力学可用于实现这一目标。这需要首先评估这种电位在表示 DPG 与其 PGI 配体和紧密结合的水分子之间的相互作用时的准确性。此类评估是通过与高级从头算量子化学 (QC) 计算进行比较来完成的。我们专注于片段从头计算 (SIBFA) 极化分子力学程序之间的相互作用总和。我们首先介绍 DPG 校准的结果。随后是关于 DPG 与主链和侧链 PGI 残基的双分子复合物的 ΔE(SIBFA) 和 ΔE(QC) 之间的比较,后者在识别位点与其结合。然后我们考虑具有越来越多的 PGI 残基的 DPG 复合物。最大的 QC 复合物包含整个识别位点,六个结构水分子总共多达 211 个原子。ΔE(SIBFA) 和 ΔE(QC) 之间可以显示出持久且令人满意的一致性。这些验证构成了对结合在 PGI 二聚体界面上的基于 DPG 的生物传感器进行大规模分子动力学模拟的重要第一步。© 2020 威利期刊公司。











































 京公网安备 11010802027423号
京公网安备 11010802027423号