当前位置:
X-MOL 学术
›
npj Syst. Biol. Appl.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mechanistic insights into bacterial metabolic reprogramming from omics-integrated genome-scale models.
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2020-01-07 , DOI: 10.1038/s41540-019-0121-4 Noushin Hadadi 1 , Vikash Pandey 2 , Anush Chiappino-Pepe 2 , Marian Morales 1 , Hector Gallart-Ayala 3 , Florence Mehl 3 , Julijana Ivanisevic 3 , Vladimir Sentchilo 1 , Jan R van der Meer 1
npj Systems Biology and Applications ( IF 3.5 ) Pub Date : 2020-01-07 , DOI: 10.1038/s41540-019-0121-4 Noushin Hadadi 1 , Vikash Pandey 2 , Anush Chiappino-Pepe 2 , Marian Morales 1 , Hector Gallart-Ayala 3 , Florence Mehl 3 , Julijana Ivanisevic 3 , Vladimir Sentchilo 1 , Jan R van der Meer 1
Affiliation
|
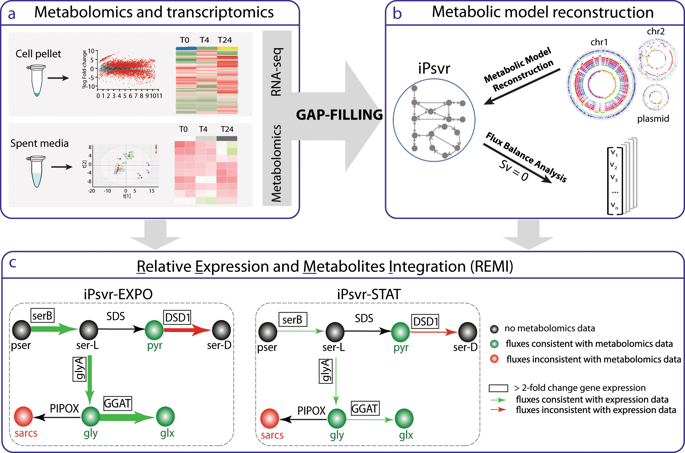
Understanding the adaptive responses of individual bacterial strains is crucial for microbiome engineering approaches that introduce new functionalities into complex microbiomes, such as xenobiotic compound metabolism for soil bioremediation. Adaptation requires metabolic reprogramming of the cell, which can be captured by multi-omics, but this data remains formidably challenging to interpret and predict. Here we present a new approach that combines genome-scale metabolic modeling with transcriptomics and exometabolomics, both of which are common tools for studying dynamic population behavior. As a realistic demonstration, we developed a genome-scale model of Pseudomonas veronii 1YdBTEX2, a candidate bioaugmentation agent for accelerated metabolism of mono-aromatic compounds in soil microbiomes, while simultaneously collecting experimental data of P. veronii metabolism during growth phase transitions. Predictions of the P. veronii growth rates and specific metabolic processes from the integrated model closely matched experimental observations. We conclude that integrative and network-based analysis can help build predictive models that accurately capture bacterial adaptation responses. Further development and testing of such models may considerably improve the successful establishment of bacterial inoculants in more complex systems.
中文翻译:

从组学整合的基因组规模模型对细菌代谢重编程的机制见解。
了解单个细菌菌株的适应性反应对于将新功能引入复杂微生物组的微生物组工程方法至关重要,例如用于土壤生物修复的外源化合物代谢。适应需要细胞的代谢重编程,这可以通过多组学捕获,但这些数据的解释和预测仍然具有巨大的挑战性。在这里,我们提出了一种新方法,将基因组规模的代谢模型与转录组学和外代谢组学相结合,这两者都是研究动态群体行为的常用工具。作为一个现实的演示,我们开发了维罗尼假单胞菌 1YdBTEX2 的基因组规模模型,这是一种候选生物增强剂,用于加速土壤微生物组中单芳香族化合物的代谢,同时收集维罗尼假单胞菌在生长阶段转变期间代谢的实验数据。综合模型对 P. veronii 生长速率和特定代谢过程的预测与实验观察结果非常吻合。我们的结论是,基于网络的综合分析可以帮助建立准确捕捉细菌适应反应的预测模型。此类模型的进一步开发和测试可能会大大提高在更复杂的系统中细菌接种剂的成功建立。
更新日期:2020-01-07
中文翻译:

从组学整合的基因组规模模型对细菌代谢重编程的机制见解。
了解单个细菌菌株的适应性反应对于将新功能引入复杂微生物组的微生物组工程方法至关重要,例如用于土壤生物修复的外源化合物代谢。适应需要细胞的代谢重编程,这可以通过多组学捕获,但这些数据的解释和预测仍然具有巨大的挑战性。在这里,我们提出了一种新方法,将基因组规模的代谢模型与转录组学和外代谢组学相结合,这两者都是研究动态群体行为的常用工具。作为一个现实的演示,我们开发了维罗尼假单胞菌 1YdBTEX2 的基因组规模模型,这是一种候选生物增强剂,用于加速土壤微生物组中单芳香族化合物的代谢,同时收集维罗尼假单胞菌在生长阶段转变期间代谢的实验数据。综合模型对 P. veronii 生长速率和特定代谢过程的预测与实验观察结果非常吻合。我们的结论是,基于网络的综合分析可以帮助建立准确捕捉细菌适应反应的预测模型。此类模型的进一步开发和测试可能会大大提高在更复杂的系统中细菌接种剂的成功建立。











































 京公网安备 11010802027423号
京公网安备 11010802027423号