当前位置:
X-MOL 学术
›
Food Control
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Rapid and Sensitive Screening and Identification of CRISPR/Cas9 Edited Rice Plants Using Quantitative Real-time PCR Coupled with High Resolution Melting Analysis
Food Control ( IF 5.6 ) Pub Date : 2020-06-01 , DOI: 10.1016/j.foodcont.2020.107088 Rong Li , Yan Ba , Yu Song , Jinjie Cui , Xiujie Zhang , Dabing Zhang , Zheng Yuan , Litao Yang
Food Control ( IF 5.6 ) Pub Date : 2020-06-01 , DOI: 10.1016/j.foodcont.2020.107088 Rong Li , Yan Ba , Yu Song , Jinjie Cui , Xiujie Zhang , Dabing Zhang , Zheng Yuan , Litao Yang
|
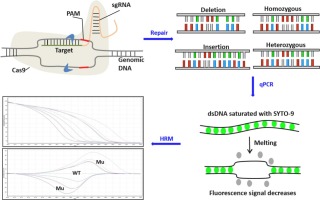
Abstract Gene-editing techniques, such as TALEN and CRISPR/Cas9, have been widely used for target DNA editing in many research fields. However, how to rapidly screen and identify the expected gene-edited products with high efficiency and low cost is still a difficult task. Here, we report the development and optimization of one such method that combines quantitative PCR with high-resolution melting analysis (qPCR-HRM) to screen and identify CRISPR/Cas9-edited rice plants. The results showed that gene-edited rice plants with small target DNA in/dels or even single base pair insertion/deletions could be successfully identified. The sensitivity of the qPCR-HRM method is as low as 1%, which is satisfying to be used for quantitative evaluation of gene-editing efficiency. Sanger sequencing results confirmed that the qPCR-HRM method also has high accuracy. It is concluded that the developed qPCR-HRM method is reproducible, accurate, and efficient for quick screening and identification of gene-edited rice plants.
中文翻译:

使用定量实时 PCR 结合高分辨率熔解分析快速、灵敏地筛选和鉴定 CRISPR/Cas9 编辑的水稻植株
摘要 TALEN 和 CRISPR/Cas9 等基因编辑技术已被广泛应用于许多研究领域的目标 DNA 编辑。然而,如何快速、高效、低成本地筛选和识别预期的基因编辑产品仍然是一项艰巨的任务。在这里,我们报告了一种这样的方法的开发和优化,该方法将定量 PCR 与高分辨率熔解分析 (qPCR-HRM) 相结合,以筛选和识别 CRISPR/Cas9 编辑的水稻植株。结果表明,基因编辑的水稻植株在/dels 中的小目标DNA 甚至单个碱基对插入/缺失都可以成功识别。qPCR-HRM 方法的灵敏度低至 1%,可满足用于基因编辑效率的定量评估。Sanger测序结果证实qPCR-HRM方法也具有较高的准确度。结论是,所开发的 qPCR-HRM 方法可重现、准确且高效,可用于基因编辑水稻植株的快速筛选和鉴定。
更新日期:2020-06-01
中文翻译:

使用定量实时 PCR 结合高分辨率熔解分析快速、灵敏地筛选和鉴定 CRISPR/Cas9 编辑的水稻植株
摘要 TALEN 和 CRISPR/Cas9 等基因编辑技术已被广泛应用于许多研究领域的目标 DNA 编辑。然而,如何快速、高效、低成本地筛选和识别预期的基因编辑产品仍然是一项艰巨的任务。在这里,我们报告了一种这样的方法的开发和优化,该方法将定量 PCR 与高分辨率熔解分析 (qPCR-HRM) 相结合,以筛选和识别 CRISPR/Cas9 编辑的水稻植株。结果表明,基因编辑的水稻植株在/dels 中的小目标DNA 甚至单个碱基对插入/缺失都可以成功识别。qPCR-HRM 方法的灵敏度低至 1%,可满足用于基因编辑效率的定量评估。Sanger测序结果证实qPCR-HRM方法也具有较高的准确度。结论是,所开发的 qPCR-HRM 方法可重现、准确且高效,可用于基因编辑水稻植株的快速筛选和鉴定。











































 京公网安备 11010802027423号
京公网安备 11010802027423号