Nature Biotechnology ( IF 33.1 ) Pub Date : 2020-01-06 , DOI: 10.1038/s41587-019-0368-8 Yanmei Dou 1 , Minseok Kwon 1 , Rachel E Rodin 2, 3, 4, 5 , Isidro Cortés-Ciriano 1, 6 , Ryan Doan 2, 3, 4 , Lovelace J Luquette 1, 7 , Alon Galor 1 , Craig Bohrson 1, 7 , Christopher A Walsh 2, 3, 4 , Peter J Park 1, 8
|
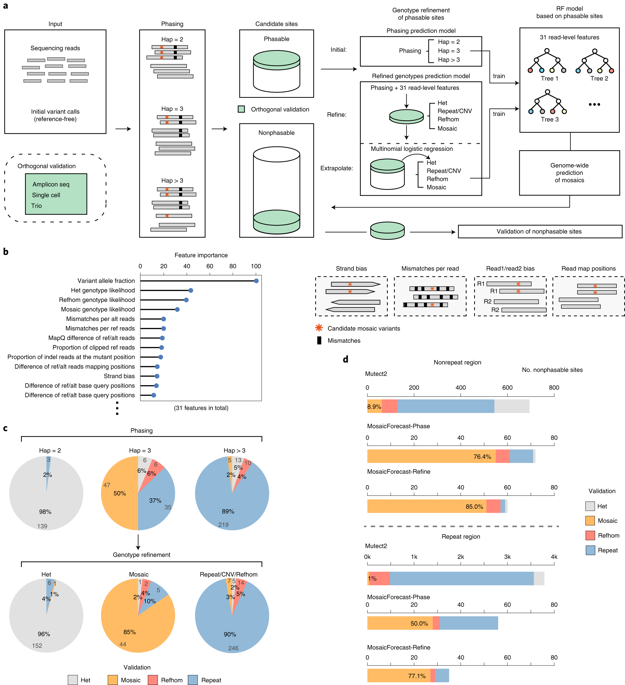
Detection of mosaic mutations that arise in normal development is challenging, as such mutations are typically present in only a minute fraction of cells and there is no clear matched control for removing germline variants and systematic artifacts. We present MosaicForecast, a machine-learning method that leverages read-based phasing and read-level features to accurately detect mosaic single-nucleotide variants and indels, achieving a multifold increase in specificity compared with existing algorithms. Using single-cell sequencing and targeted sequencing, we validated 80–90% of the mosaic single-nucleotide variants and 60–80% of indels detected in human brain whole-genome sequencing data. Our method should help elucidate the contribution of mosaic somatic mutations to the origin and development of disease.
中文翻译:

在没有匹配对照的情况下准确检测测序数据中的镶嵌变异
检测正常发育中出现的镶嵌突变具有挑战性,因为此类突变通常仅存在于一小部分细胞中,并且没有明确的匹配对照来去除种系变异和系统伪影。我们提出了 MosaicForecast,这是一种机器学习方法,它利用基于读取的定相和读取级特征来准确检测镶嵌单核苷酸变异和插入缺失,与现有算法相比,特异性提高了多倍。使用单细胞测序和靶向测序,我们验证了人脑全基因组测序数据中检测到的 80-90% 的镶嵌单核苷酸变体和 60-80% 的插入缺失。我们的方法应该有助于阐明嵌合体细胞突变对疾病起源和发展的贡献。











































 京公网安备 11010802027423号
京公网安备 11010802027423号