当前位置:
X-MOL 学术
›
ACS Appl. Nano Mater.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Molecular Dynamics Simulation Study of Transverse and Longitudinal Ionic Currents in Solid-State Nanopore DNA Sequencing
ACS Applied Nano Materials ( IF 5.3 ) Pub Date : 2020-01-16 , DOI: 10.1021/acsanm.9b02280 Mohsen Farshad 1 , Jayendran C. Rasaiah 1
ACS Applied Nano Materials ( IF 5.3 ) Pub Date : 2020-01-16 , DOI: 10.1021/acsanm.9b02280 Mohsen Farshad 1 , Jayendran C. Rasaiah 1
Affiliation
|
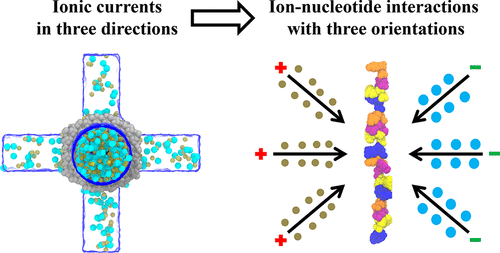
We report all-atom molecular dynamics (MD) simulations of single-stranded DNA (ssDNA) translocation in 1 M KCl solution through a silicon nitride solid-state nanopore with one or two nanochannels perpendicular to the nanopore. We measure the longitudinal and transverse ionic currents generated through the pores under voltage biases applied longitudinally and transversely across the pores. During fast translocation of homo-oligonucleotides through the pore, the characteristic signals of nucleotides resulting from ion–nucleotide interactions cannot be distinguished. These signals are buried in fluctuations of the ions caused by thermal energy at high sampling frequency. A pattern recognition neural network shows that the averaged transverse and longitudinal ionic currents and their combination enable the canonical A, G, T, and C nucleotide classifications to be recognized with an accuracy of 81.4%. Further improvements can be explored with machine learning algorithms, larger databases, and slower translocation rates at lower voltages biases that would require greater computer resources.
中文翻译:

固态纳米孔DNA测序中横向和纵向离子流的分子动力学模拟研究
我们报告了单链DNA(ssDNA)在1 M KCl溶液中穿过具有一个或两个垂直于纳米孔的纳米通道的氮化硅固态纳米孔的全原子分子动力学(MD)模拟。我们测量在纵向和横向施加在孔隙上的电压偏置下,通过孔隙产生的纵向和横向离子电流。在同型寡核苷酸通过孔快速移位的过程中,无法区分由离子-核苷酸相互作用产生的核苷酸特征信号。这些信号掩埋在高采样频率下由热能引起的离子波动中。模式识别神经网络表明,平均的横向和纵向离子流及其组合使规范的A,G,T,和C核苷酸分类的准确度为81.4%。可以使用机器学习算法,更大的数据库以及更低的电压偏置(需要更多的计算机资源)下的较慢移位速率来探索进一步的改进。
更新日期:2020-01-16
中文翻译:

固态纳米孔DNA测序中横向和纵向离子流的分子动力学模拟研究
我们报告了单链DNA(ssDNA)在1 M KCl溶液中穿过具有一个或两个垂直于纳米孔的纳米通道的氮化硅固态纳米孔的全原子分子动力学(MD)模拟。我们测量在纵向和横向施加在孔隙上的电压偏置下,通过孔隙产生的纵向和横向离子电流。在同型寡核苷酸通过孔快速移位的过程中,无法区分由离子-核苷酸相互作用产生的核苷酸特征信号。这些信号掩埋在高采样频率下由热能引起的离子波动中。模式识别神经网络表明,平均的横向和纵向离子流及其组合使规范的A,G,T,和C核苷酸分类的准确度为81.4%。可以使用机器学习算法,更大的数据库以及更低的电压偏置(需要更多的计算机资源)下的较慢移位速率来探索进一步的改进。









































 京公网安备 11010802027423号
京公网安备 11010802027423号