Protein & Peptide Letters ( IF 1.0 ) Pub Date : 2020-05-01 , DOI: 10.2174/0929866526666191028162302 Hua Wan 1 , Jian-Ming Li 1 , Huang Ding 1 , Shuo-Xin Lin 2 , Shu-Qin Tu 1 , Xu-Hong Tian 1 , Jian-Ping Hu 3 , Shan Chang 4
|
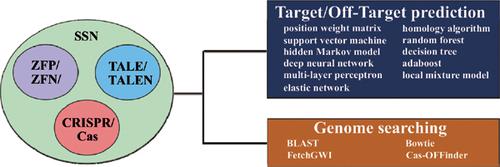
Understanding the interaction mechanism of proteins and nucleic acids is one of the most fundamental problems for genome editing with engineered nucleases. Due to some limitations of experimental investigations, computational methods have played an important role in obtaining the knowledge of protein-nucleic acid interaction. Over the past few years, dozens of computational tools have been used for identification of nucleic acid binding site for site-specific proteins and design of site-specific nucleases because of their significant advantages in genome editing. Here, we review existing widely-used computational tools for target prediction of site-specific proteins as well as off-target prediction of site-specific nucleases. This article provides a list of on-line prediction tools according to their features followed by the description of computational methods used by these tools, which range from various sequence mapping algorithms (like Bowtie, FetchGWI and BLAST) to different machine learning methods (such as Support Vector Machine, hidden Markov models, Random Forest, elastic network and deep neural networks). We also make suggestions on the further development in improving the accuracy of prediction methods. This survey will provide a reference guide for computational biologists working in the field of genome editing.
中文翻译:

用于位点特异性蛋白质和核酸酶的核酸结合位点预测的计算工具概述。
了解蛋白质和核酸的相互作用机制是利用工程核酸酶进行基因组编辑的最基本问题之一。由于实验研究的某些局限性,计算方法在获得蛋白质-核酸相互作用的知识方面发挥了重要作用。在过去的几年中,由于它们在基因组编辑方面的显着优势,数十种计算工具已用于鉴定位点特异性蛋白质的核酸结合位点和位点特异性核酸酶的设计。在这里,我们回顾了用于位点特异性蛋白质的目标预测以及位点特异性核酸酶的脱靶预测的现有广泛使用的计算工具。本文根据其功能提供了一系列在线预测工具,然后介绍了这些工具使用的计算方法,这些方法的范围从各种序列映射算法(例如Bowtie,FetchGWI和BLAST)到不同的机器学习方法(例如支持向量机,隐马尔可夫模型,随机森林,弹性网络和深度神经网络)。我们还为提高预测方法的准确性提出了进一步的建议。这项调查将为从事基因组编辑领域的计算生物学家提供参考指南。弹性网络和深度神经网络)。我们还为提高预测方法的准确性提出了进一步的建议。该调查将为基因组编辑领域的计算生物学家提供参考指南。弹性网络和深度神经网络)。我们还为提高预测方法的准确性提出了进一步的建议。该调查将为基因组编辑领域的计算生物学家提供参考指南。











































 京公网安备 11010802027423号
京公网安备 11010802027423号