Current Computer-Aided Drug Design ( IF 1.5 ) Pub Date : 2020-11-30 , DOI: 10.2174/1573409915666190918150136 Omar Husham Ahmed Al-Attraqchi 1 , Katharigatta N Venugopala 2
|
Background: Glutaminyl Cyclase (QC) is a novel target in the battle against Alzheimer’s disease, a highly prevalent neurodegenerative disorder. QC inhibitors have the potential to be developed as therapeutically useful anti-Alzheimer’s disease agents.
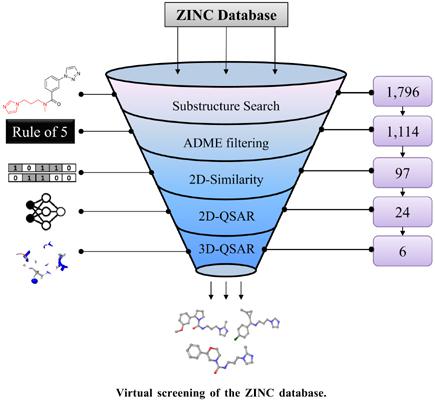
Methods: Linear and non-linear 2D-Quantitative Structure-Activity Relationship (QSAR) models were developed using Stepwise Multiple Linear Regression (S-MLR) and neural networks. Partial least squares (PLS) method was used to develop a 3D-QSAR model. Also, the developed models were used in virtual screening of the ZINC database to identify potential QC inhibitors.
Results: The 2D neural network model showed superior predictive ability, as demonstrated by the validation parameters R2 = 0.933, Q2 = 0.886 and R2 pred = 0.911. The 3D-QSAR model’s steric and electrostatic fields’ isocontour maps were visualized and revealed important structural requirements associated with good activity. The virtual screening identified six compounds as potentially active QC inhibitors with improved pharmacokinetic profiles.
Conclusion: The developed QSAR models can be used to predict the activity of compounds not yet synthesized and prioritized for their synthesis and biological evaluation. Also, potentially active QC inhibitors have been identified with attractive lead-like properties that can be used to develop anti- Alzheimer’s disease agents.
中文翻译:

基于咪唑的谷氨酰胺酰环化酶抑制剂的 2D 和 3D-QSAR 建模。
背景:谷氨酰胺酰环化酶 (QC) 是对抗阿尔茨海默病(一种高度流行的神经退行性疾病)的新靶点。QC 抑制剂有可能被开发为治疗上有用的抗阿尔茨海默病药物。
方法:使用逐步多元线性回归 (S-MLR) 和神经网络开发线性和非线性 2D 定量结构-活动关系 (QSAR) 模型。偏最小二乘 (PLS) 方法用于开发 3D-QSAR 模型。此外,开发的模型用于 ZINC 数据库的虚拟筛选,以识别潜在的 QC 抑制剂。
结果:二维神经网络模型显示出卓越的预测能力,验证参数 R 2 = 0.933、Q 2 = 0.886 和 R 2 pred = 0.911证明了这一点。3D-QSAR 模型的立体和静电场的等值线图被可视化,并揭示了与良好活动相关的重要结构要求。虚拟筛选确定了六种化合物作为具有改进药代动力学特征的潜在活性 QC 抑制剂。
结论:开发的 QSAR 模型可用于预测尚未合成的化合物的活性,并对其合成和生物学评估进行优先排序。此外,已鉴定出具有有吸引力的铅样特性的潜在活性 QC 抑制剂,可用于开发抗阿尔茨海默病药物。











































 京公网安备 11010802027423号
京公网安备 11010802027423号