Current Computer-Aided Drug Design ( IF 1.5 ) Pub Date : 2020-07-31 , DOI: 10.2174/1573409915666190617165643 Hanane Boucherit 1 , Abdelouahab Chikhi 1 , Abderrahmane Bensegueni 1 , Amina Merzoug 1 , Jean-Michel Bolla 2
|
Background: The great emergence of multi-resistant bacterial strains and the low renewal of antibiotics molecules are leading human and veterinary medicine to certain therapeutic impasses. Therefore, there is an urgent need to find new therapeutic alternatives including new molecules in the current treatments of infectious diseases. Methionine aminopeptidase (MetAP) is a promising target for developing new antibiotics because it is essential for bacterial survival.
Objective: To screen for potential MetAP inhibitors by in silico virtual screening of the ZINC database and evaluate the best potential lead molecules by in vitro studies.
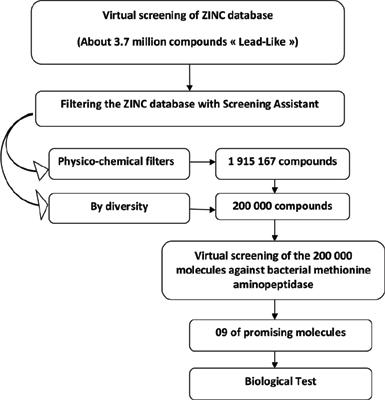
Methods: We have considered 200,000 compounds from the ZINC database for virtual screening with FlexX software to identify potential inhibitors against bacterial MetAP. Nine chemical compounds of the top hits predicted were purchased and evaluated in vitro. The antimicrobial activity of each inhibitor of MetAP was tested by the disc-diffusion assay against one Gram-positive (Staphylococcus aureus) and two Gram-negative (Escherichia coli & Pseudomonas aeruginosa) bacteria. Among the studied compounds, compounds ZINC04785369 and ZINC03307916 showed promising antibacterial activity. To further characterize their efficacy, the minimum inhibitory concentration was determined for each compound by the microdilution method which showed significant results.
Results: These results suggest compounds ZINC04785369 and ZINC03307916 as promising molecules for developing MetAP inhibitors.
Conclusion: Furthermore, they could therefore serve as lead molecules for further chemical modifications to obtain clinically useful antibacterial agents.
中文翻译:

通过基于结构的ZINC数据库虚拟筛选方法和体外验证研究新型细菌蛋氨酸氨基肽酶抑制剂。
背景:多重耐药菌的大量涌现和抗生素分子的低更新正导致人类和兽医医学陷入某些治疗僵局。因此,迫切需要在当前的传染病治疗中寻找包括新分子在内的新的治疗替代方法。蛋氨酸氨基肽酶(MetAP)是开发新抗生素的有希望的靶标,因为它对细菌存活至关重要。
目的:通过对ZINC数据库的计算机虚拟筛选来筛选潜在的MetAP抑制剂,并通过体外研究评估最佳的潜在先导分子。
方法:我们已经考虑了ZINC数据库中的20万种化合物,以用FlexX软件进行虚拟筛选,以鉴定潜在的抗细菌MetAP抑制剂。购买了9种预测最高命中的化合物,并在体外进行了评估。通过碟片扩散测定法测试了每种MetAP抑制剂对一种革兰氏阳性菌(金黄色葡萄球菌)和两种革兰氏阴性菌(大肠杆菌和铜绿假单胞菌)的抗菌活性。在研究的化合物中,化合物ZINC04785369和ZINC03307916显示出令人鼓舞的抗菌活性。为了进一步表征其功效,通过微稀释法确定了每种化合物的最低抑菌浓度,结果显示了显着的效果。
结果:这些结果表明,化合物ZINC04785369和ZINC03307916是开发MetAP抑制剂的有前途的分子。
结论:此外,它们可以用作先导分子,用于进一步的化学修饰以获得临床上有用的抗菌剂。











































 京公网安备 11010802027423号
京公网安备 11010802027423号