Current Computer-Aided Drug Design ( IF 1.5 ) Pub Date : 2020-07-31 , DOI: 10.2174/1573409915666190708103132 Fortunatus C Ezebuo 1 , Ikemefuna C Uzochukwu 1
|
Background: Sulfotransferase family comprises key enzymes involved in drug metabolism. Oxamniquine is a pro-drug converted into its active form by schistosomal sulfotransferase. The conformational dynamics of side-chain amino acid residues at the binding site of schistosomal sulfotransferase towards activation of oxamniquine has not received attention.
Objective: The study investigated the conformational dynamics of binding site residues in free and oxamniquine bound schistosomal sulfotransferase systems and their contribution to the mechanism of oxamniquine activation by schistosomal sulfotransferase using molecular dynamics simulations and binding energy calculations.
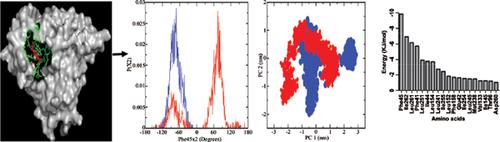
Methods: Schistosomal sulfotransferase was obtained from Protein Data Bank and both the free and oxamniquine bound forms were subjected to molecular dynamics simulations using GROMACS-4.5.5 after modeling it’s missing amino acid residues with SWISS-MODEL. Amino acid residues at its binding site for oxamniquine was determined and used for Principal Component Analysis and calculations of side-chain dihedrals. In addition, binding energy of the oxamniquine bound system was calculated using g_MMPBSA.
Results: The results showed that binding site amino acid residues in free and oxamniquine bound sulfotransferase sampled different conformational space involving several rotameric states. Importantly, Phe45, Ile145 and Leu241 generated newly induced conformations, whereas Phe41 exhibited shift in equilibrium of its conformational distribution. In addition, the result showed binding energy of -130.091 ± 8.800 KJ/mol and Phe45 contributed -9.8576 KJ/mol.
Conclusion: The results showed that schistosomal sulfotransferase binds oxamniquine by relying on hybrid mechanism of induced fit and conformational selection models. The findings offer new insight into sulfotransferase engineering and design of new drugs that target sulfotransferase.
中文翻译:

血吸虫硫转移酶与奥沙尼喹的相互作用涉及诱导拟合和构象选择的混合机制。
背景:磺基转移酶家族包含参与药物代谢的关键酶。Oxamniquine是一种通过血吸虫磺基转移酶转化为其活性形式的前药。在血吸虫磺基转移酶的结合位点上的侧链氨基酸残基的构象动力学未引起人们的注意。
目的:通过分子动力学模拟和结合能计算,研究游离和奥沙米喹结合的血吸虫磺基转移酶系统中结合位点残基的构象动力学及其对血吸虫磺基转移酶激活氧苯甲喹机制的贡献。
方法:从蛋白质数据库中获得血吸虫磺基转移酶,并用SWISS-MODEL对缺失的氨基酸残基进行建模,然后使用GROMACS-4.5.5对游离形式和奥沙米醌结合形式进行分子动力学模拟。测定了其与氧杂喹啉结合位点的氨基酸残基,并将其用于主成分分析和侧链二面体的计算。另外,使用g_MMPBSA计算氧杂苯醌结合系统的结合能。
结果:结果表明,游离的和氧杂苯甲酰胺结合的磺基转移酶的结合位点氨基酸残基取样了涉及几种旋转异构状态的不同构象空间。重要的是,Phe45,Ile145和Leu241产生新诱导的构象,而Phe41在其构象分布的平衡中表现出位移。另外,结果显示结合能为-130.091±8.800KJ / mol,Phe45贡献为-9.8576KJ / mol。
结论:结果表明血吸虫磺基转移酶依赖于诱导拟合和构象选择模型的杂合机制结合氧肟酰胺。这些发现为磺基转移酶工程设计和针对磺基转移酶的新药设计提供了新的见解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号