Protein & Peptide Letters ( IF 1.6 ) Pub Date : 2020-05-01 , DOI: 10.2174/0929866526666190410124110 Yijie Ding 1 , Jijun Tang 2, 3 , Fei Guo 3
|
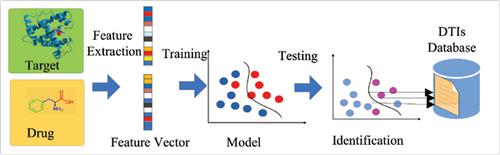
The identification of Drug-Target Interactions (DTIs) is an important process in drug discovery and medical research. However, the tradition experimental methods for DTIs identification are still time consuming, extremely expensive and challenging. In the past ten years, various computational methods have been developed to identify potential DTIs. In this paper, the identification methods of DTIs are summarized. What's more, several state-of-the-art computational methods are mainly introduced, containing network-based method and machine learning-based method. In particular, for machine learning-based methods, including the supervised and semisupervised models, have essential differences in the approach of negative samples. Although these effective computational models in identification of DTIs have achieved significant improvements, network-based and machine learning-based methods have their disadvantages, respectively. These computational methods are evaluated on four benchmark data sets via values of Area Under the Precision Recall curve (AUPR).
中文翻译:

药物-靶标相互作用预测的计算模型。
药物-靶标相互作用(DTI)的识别是药物发现和医学研究中的重要过程。但是,用于DTI识别的传统实验方法仍然很耗时,极其昂贵且具有挑战性。在过去的十年中,已经开发出各种计算方法来识别潜在的DTI。本文总结了DTI的识别方法。此外,主要介绍了几种最新的计算方法,包括基于网络的方法和基于机器学习的方法。尤其是,对于基于机器学习的方法,包括监督模型和半监督模型,在否定样本方法上存在本质差异。尽管这些用于识别DTI的有效计算模型已经取得了重大进步,基于网络的方法和基于机器学习的方法分别具有缺点。这些计算方法通过“精确召回曲线下的面积”(AUPR)的值在四个基准数据集上进行评估。



























 京公网安备 11010802027423号
京公网安备 11010802027423号