Nature Methods ( IF 36.1 ) Pub Date : 2018-10-30 , DOI: 10.1038/s41592-018-0176-y Eric A. Franzosa , Lauren J. McIver , Gholamali Rahnavard , Luke R. Thompson , Melanie Schirmer , George Weingart , Karen Schwarzberg Lipson , Rob Knight , J. Gregory Caporaso , Nicola Segata , Curtis Huttenhower
|
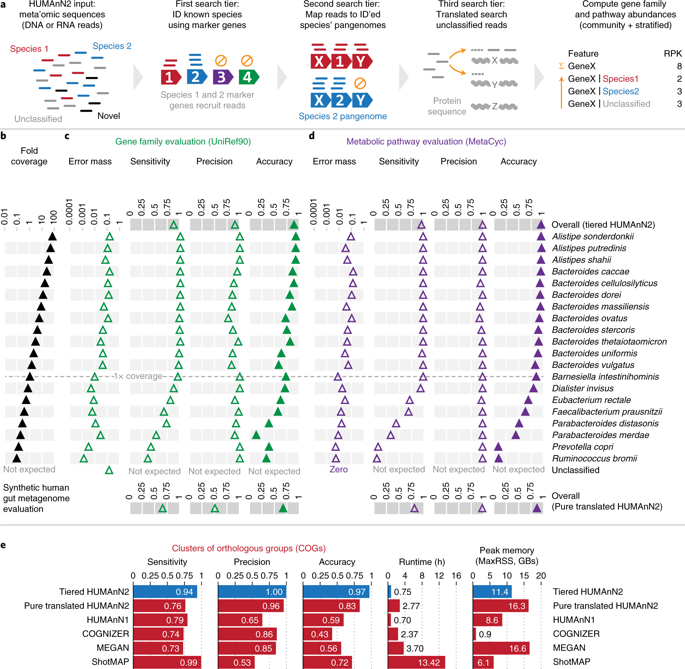
Functional profiles of microbial communities are typically generated using comprehensive metagenomic or metatranscriptomic sequence read searches, which are time-consuming, prone to spurious mapping, and often limited to community-level quantification. We developed HUMAnN2, a tiered search strategy that enables fast, accurate, and species-resolved functional profiling of host-associated and environmental communities. HUMAnN2 identifies a community’s known species, aligns reads to their pangenomes, performs translated search on unclassified reads, and finally quantifies gene families and pathways. Relative to pure translated search, HUMAnN2 is faster and produces more accurate gene family profiles. We applied HUMAnN2 to study clinal variation in marine metabolism, ecological contribution patterns among human microbiome pathways, variation in species’ genomic versus transcriptional contributions, and strain profiling. Further, we introduce ‘contributional diversity’ to explain patterns of ecological assembly across different microbial community types.
中文翻译:

元基因组和元转录组的物种级功能分析
微生物群落的功能概况通常是使用全面的宏基因组学或元转录组序列阅读搜索生成的,这些过程很耗时,容易进行虚假作图,并且通常仅限于群落一级的定量分析。我们开发了HUMAnN2,这是一种分层的搜索策略,可对与宿主相关的社区和环境社区进行快速,准确和物种解析的功能分析。HUMAnN2可以识别一个社区的已知物种,将读物与其泛基因组对齐,对未分类的读物进行翻译搜索,最后量化基因家族和途径。相对于纯翻译搜索,HUMAnN2更快,并且产生更准确的基因家族谱。我们使用HUMAnN2研究了海洋代谢中的临床变化,人类微生物组途径之间的生态贡献模式,物种基因组与转录贡献的差异以及菌株概况分析。此外,我们引入“贡献多样性”来解释跨不同微生物群落类型的生态装配模式。











































 京公网安备 11010802027423号
京公网安备 11010802027423号