Nature Methods ( IF 36.1 ) Pub Date : 2018-11-30 , DOI: 10.1038/s41592-018-0213-x Shijie C Zheng 1 , Charles E Breeze 2 , Stephan Beck 3 , Andrew E Teschendorff 1, 3, 4
|
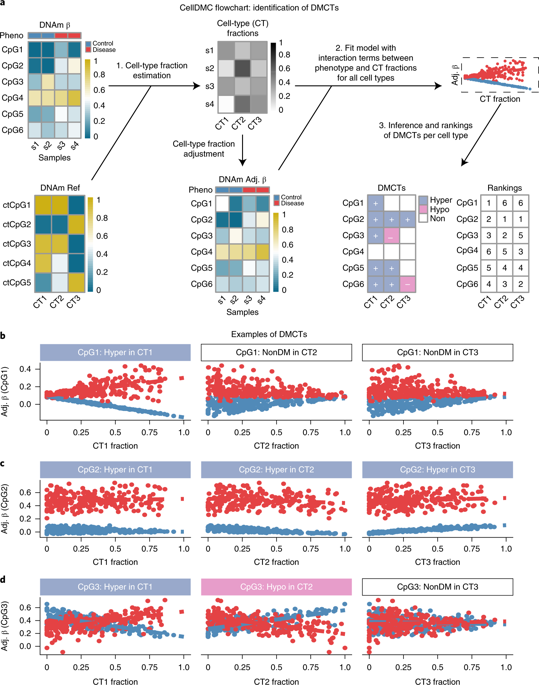
An outstanding challenge of epigenome-wide association studies (EWASs) performed in complex tissues is the identification of the specific cell type(s) responsible for the observed differential DNA methylation. Here we present a statistical algorithm called CellDMC (https://github.com/sjczheng/EpiDISH), which can identify differentially methylated positions and the specific cell type(s) driving the differential methylation. We validated CellDMC on in silico mixtures of DNA methylation data generated with different technologies, as well as on real mixtures from epigenome-wide association and cancer epigenome studies. CellDMC achieved over 90% sensitivity and specificity in scenarios where current state-of-the-art methods did not identify differential methylation. By applying CellDMC to an EWAS performed in buccal swabs, we identified smoking-associated differentially methylated positions occurring in the epithelial compartment, which we validated in smoking-related lung cancer. CellDMC may be useful in the identification of causal DNA-methylation alterations in disease.
中文翻译:

在表观基因组范围的关联研究中鉴定差异甲基化细胞类型。
在复杂组织中进行的表观基因组关联研究(EWAS)的一大挑战是鉴定负责观察到的差异DNA甲基化的特定细胞类型。在这里,我们介绍了一种称为CellDMC(https://github.com/sjczheng/EpiDISH)的统计算法,该算法可以识别差异甲基化的位置和驱动差异甲基化的特定细胞类型。我们使用不同技术生成的DNA甲基化数据的计算机模拟混合物以及表观基因组范围内的关联和癌症表观基因组研究的真实混合物对CellDMC进行了验证。在当前最先进的方法无法识别差异甲基化的情况下,CellDMC达到了90%以上的灵敏度和特异性。通过将CellDMC应用于在颊拭子中进行的EWAS,我们确定了与吸烟相关的甲基化差异发生在上皮区室中,我们已在吸烟相关的肺癌中进行了验证。CellDMC可用于鉴定疾病中的因果DNA甲基化改变。











































 京公网安备 11010802027423号
京公网安备 11010802027423号