当前位置:
X-MOL 学术
›
Nat. Commun.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Discovering de novo peptide substrates for enzymes using machine learning.
Nature Communications ( IF 14.7 ) Pub Date : 2018-12-07 , DOI: 10.1038/s41467-018-07717-6 Lorillee Tallorin , JiaLei Wang , Woojoo E. Kim , Swagat Sahu , Nicolas M. Kosa , Pu Yang , Matthew Thompson , Michael K. Gilson , Peter I. Frazier , Michael D. Burkart , Nathan C. Gianneschi
Nature Communications ( IF 14.7 ) Pub Date : 2018-12-07 , DOI: 10.1038/s41467-018-07717-6 Lorillee Tallorin , JiaLei Wang , Woojoo E. Kim , Swagat Sahu , Nicolas M. Kosa , Pu Yang , Matthew Thompson , Michael K. Gilson , Peter I. Frazier , Michael D. Burkart , Nathan C. Gianneschi
|
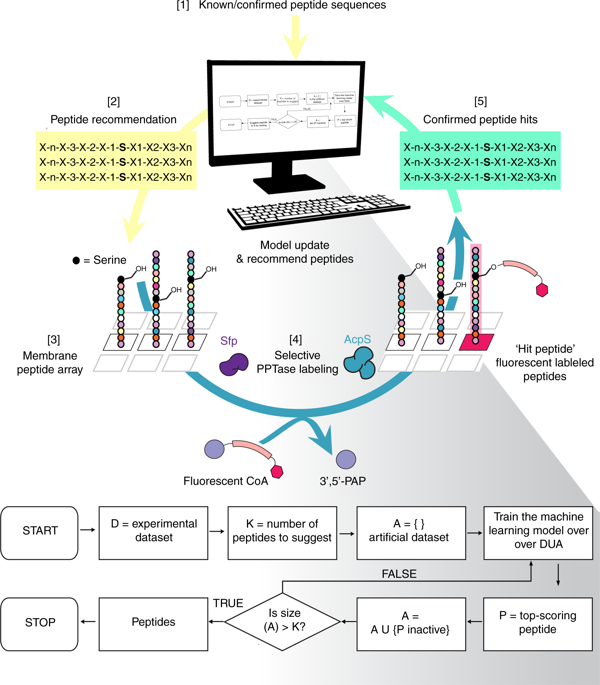
The discovery of peptide substrates for enzymes with exclusive, selective activities is a central goal in chemical biology. In this paper, we develop a hybrid computational and biochemical method to rapidly optimize peptides for specific, orthogonal biochemical functions. The method is an iterative machine learning process by which experimental data is deposited into a mathematical algorithm that selects potential peptide substrates to be tested experimentally. Once tested, the algorithm uses the experimental data to refine future selections. This process is repeated until a suitable set of de novo peptide substrates are discovered. We employed this technology to discover orthogonal peptide substrates for 4'-phosphopantetheinyl transferase, an enzyme class that covalently modifies proteins. In this manner, we have demonstrated that machine learning can be leveraged to guide peptide optimization for specific biochemical functions not immediately accessible by biological screening techniques, such as phage display and random mutagenesis.
中文翻译:

使用机器学习发现酶的从头肽底物。
发现具有排他性,选择性活性的酶的肽底物是化学生物学的主要目标。在本文中,我们开发了一种混合计算和生化方法,可快速优化针对特定正交生化功能的肽。该方法是一个迭代的机器学习过程,通过该过程,将实验数据沉积到数学算法中,该数学算法选择要进行实验测试的潜在肽底物。测试完成后,该算法将使用实验数据来完善将来的选择。重复该过程,直到发现合适的一组从头肽底物。我们采用了这项技术来发现4'-磷酸邻苯二甲酰转移酶(一种共价修饰蛋白质的酶类)的正交肽底物。以这种方式,
更新日期:2018-12-07
中文翻译:

使用机器学习发现酶的从头肽底物。
发现具有排他性,选择性活性的酶的肽底物是化学生物学的主要目标。在本文中,我们开发了一种混合计算和生化方法,可快速优化针对特定正交生化功能的肽。该方法是一个迭代的机器学习过程,通过该过程,将实验数据沉积到数学算法中,该数学算法选择要进行实验测试的潜在肽底物。测试完成后,该算法将使用实验数据来完善将来的选择。重复该过程,直到发现合适的一组从头肽底物。我们采用了这项技术来发现4'-磷酸邻苯二甲酰转移酶(一种共价修饰蛋白质的酶类)的正交肽底物。以这种方式,











































 京公网安备 11010802027423号
京公网安备 11010802027423号