Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Controlling orthogonal ribosome subunit interactions enables evolution of new function
Nature ( IF 50.5 ) Pub Date : 2018-12-01 , DOI: 10.1038/s41586-018-0773-z Wolfgang H Schmied 1 , Zakir Tnimov 1 , Chayasith Uttamapinant 1, 2 , Christopher D Rae 1 , Stephen D Fried 1, 3 , Jason W Chin 1
Nature ( IF 50.5 ) Pub Date : 2018-12-01 , DOI: 10.1038/s41586-018-0773-z Wolfgang H Schmied 1 , Zakir Tnimov 1 , Chayasith Uttamapinant 1, 2 , Christopher D Rae 1 , Stephen D Fried 1, 3 , Jason W Chin 1
Affiliation
|
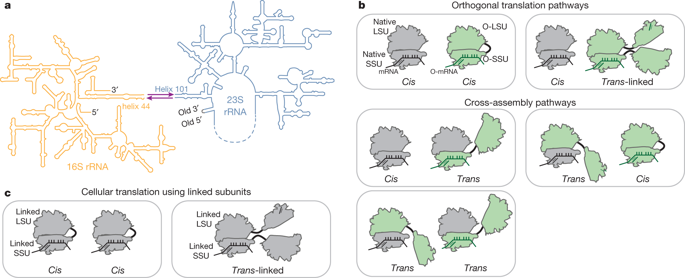
Orthogonal ribosomes are unnatural ribosomes that are directed towards orthogonal messenger RNAs in Escherichia coli, through an altered version of the 16S ribosomal RNA of the small subunit1. Directed evolution of orthogonal ribosomes has provided access to new ribosomal function, and the evolved orthogonal ribosomes have enabled the encoding of multiple non-canonical amino acids into proteins2–4. The original orthogonal ribosomes shared the pool of 23S ribosomal RNAs, contained in the large subunit, with endogenous ribosomes. Selectively directing a new 23S rRNA to an orthogonal mRNA, by controlling the association between the orthogonal 16S rRNAs and 23S rRNAs, would enable the evolution of new function in the large subunit. Previous work covalently linked orthogonal 16S rRNA and a circularly permuted 23S rRNA to create orthogonal ribosomes with low activity5,6; however, the linked subunits in these ribosomes do not associate specifically with each other, and mediate translation by associating with endogenous subunits. Here we discover engineered orthogonal ‘stapled’ ribosomes (with subunits linked through an optimized RNA staple) with activities comparable to that of the parent orthogonal ribosome; they minimize association with endogenous subunits and mediate translation of orthogonal mRNAs through the association of stapled subunits. We evolve cells with genomically encoded stapled ribosomes as the sole ribosomes, which support cellular growth at similar rates to natural ribosomes. Moreover, we visualize the engineered stapled ribosome structure by cryo-electron microscopy at 3.0 Å, revealing how the staple links the subunits and controls their association. We demonstrate the utility of controlling subunit association by evolving orthogonal stapled ribosomes which efficiently polymerize a sequence of monomers that the natural ribosome is intrinsically unable to translate. Our work provides a foundation for evolving the rRNA of the entire orthogonal ribosome for the encoded cellular synthesis of non-canonical biological polymers7.Orthogonal ribosomes are engineered in which the two subunits are stapled together in a way that limits association with endogenous subunits in cells, enabling the evolution of new functionality in the orthogonal ribosome.
中文翻译:

控制正交核糖体亚基相互作用可以实现新功能的进化
正交核糖体是非天然核糖体,通过小亚基 16S 核糖体 RNA 的改变版本,定向至大肠杆菌中的正交信使 RNA。正交核糖体的定向进化提供了获得新核糖体功能的途径,并且进化的正交核糖体能够将多种非规范氨基酸编码成蛋白质2-4。原始正交核糖体与内源核糖体共享大亚基中包含的 23S 核糖体 RNA 库。通过控制正交16S rRNA和23S rRNA之间的关联,选择性地将新的23S rRNA导向正交mRNA,将使大亚基中新功能的进化成为可能。之前的工作共价连接正交 16S rRNA 和循环排列的 23S rRNA,以创建低活性的正交核糖体 5,6;然而,这些核糖体中连接的亚基彼此之间并不特异性结合,而是通过与内源亚基结合来介导翻译。在这里,我们发现了工程正交“钉合”核糖体(亚基通过优化的 RNA 钉合连接),其活性与亲本正交核糖体相当;它们最大限度地减少与内源亚基的关联,并通过钉合亚基的关联介导正交 mRNA 的翻译。我们用基因组编码的钉合核糖体作为唯一的核糖体来进化细胞,它以与天然核糖体相似的速率支持细胞生长。此外,我们通过冷冻电子显微镜在 3.0 Å 下可视化了工程化的钉合核糖体结构,揭示了钉合如何连接亚基并控制它们的关联。 我们证明了通过进化正交钉合核糖体来控制亚基关联的效用,该核糖体有效地聚合了天然核糖体本质上无法翻译的一系列单体。我们的工作为进化整个正交核糖体的 rRNA 以编码细胞合成非典型生物聚合物奠定了基础7。正交核糖体经过工程设计,其中两个亚基以限制与细胞内源亚基关联的方式钉在一起,使正交核糖体中新功能的进化成为可能。
更新日期:2018-12-01
中文翻译:

控制正交核糖体亚基相互作用可以实现新功能的进化
正交核糖体是非天然核糖体,通过小亚基 16S 核糖体 RNA 的改变版本,定向至大肠杆菌中的正交信使 RNA。正交核糖体的定向进化提供了获得新核糖体功能的途径,并且进化的正交核糖体能够将多种非规范氨基酸编码成蛋白质2-4。原始正交核糖体与内源核糖体共享大亚基中包含的 23S 核糖体 RNA 库。通过控制正交16S rRNA和23S rRNA之间的关联,选择性地将新的23S rRNA导向正交mRNA,将使大亚基中新功能的进化成为可能。之前的工作共价连接正交 16S rRNA 和循环排列的 23S rRNA,以创建低活性的正交核糖体 5,6;然而,这些核糖体中连接的亚基彼此之间并不特异性结合,而是通过与内源亚基结合来介导翻译。在这里,我们发现了工程正交“钉合”核糖体(亚基通过优化的 RNA 钉合连接),其活性与亲本正交核糖体相当;它们最大限度地减少与内源亚基的关联,并通过钉合亚基的关联介导正交 mRNA 的翻译。我们用基因组编码的钉合核糖体作为唯一的核糖体来进化细胞,它以与天然核糖体相似的速率支持细胞生长。此外,我们通过冷冻电子显微镜在 3.0 Å 下可视化了工程化的钉合核糖体结构,揭示了钉合如何连接亚基并控制它们的关联。 我们证明了通过进化正交钉合核糖体来控制亚基关联的效用,该核糖体有效地聚合了天然核糖体本质上无法翻译的一系列单体。我们的工作为进化整个正交核糖体的 rRNA 以编码细胞合成非典型生物聚合物奠定了基础7。正交核糖体经过工程设计,其中两个亚基以限制与细胞内源亚基关联的方式钉在一起,使正交核糖体中新功能的进化成为可能。











































 京公网安备 11010802027423号
京公网安备 11010802027423号