Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Single-cell mapping of lineage and identity in direct reprogramming
Nature ( IF 50.5 ) Pub Date : 2018-12-01 , DOI: 10.1038/s41586-018-0744-4 Brent A Biddy 1, 2, 3 , Wenjun Kong 1, 2, 3 , Kenji Kamimoto 1, 2, 3 , Chuner Guo 1, 2, 3 , Sarah E Waye 1, 2, 3 , Tao Sun 1, 2, 3, 4 , Samantha A Morris 1, 2, 3
Nature ( IF 50.5 ) Pub Date : 2018-12-01 , DOI: 10.1038/s41586-018-0744-4 Brent A Biddy 1, 2, 3 , Wenjun Kong 1, 2, 3 , Kenji Kamimoto 1, 2, 3 , Chuner Guo 1, 2, 3 , Sarah E Waye 1, 2, 3 , Tao Sun 1, 2, 3, 4 , Samantha A Morris 1, 2, 3
Affiliation
|
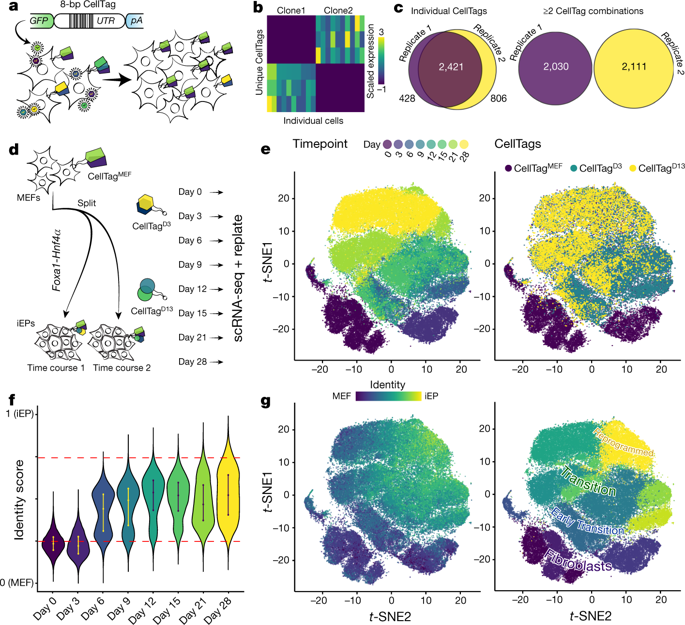
Direct lineage reprogramming involves the conversion of cellular identity. Single-cell technologies are useful for deconstructing the considerable heterogeneity that emerges during lineage conversion. However, lineage relationships are typically lost during cell processing, complicating trajectory reconstruction. Here we present ‘CellTagging’, a combinatorial cell-indexing methodology that enables parallel capture of clonal history and cell identity, in which sequential rounds of cell labelling enable the construction of multi-level lineage trees. CellTagging and longitudinal tracking of fibroblast to induced endoderm progenitor reprogramming reveals two distinct trajectories: one leading to successfully reprogrammed cells, and one leading to a ‘dead-end’ state, paths determined in the earliest stages of lineage conversion. We find that expression of a putative methyltransferase, Mettl7a1, is associated with the successful reprogramming trajectory; adding Mettl7a1 to the reprogramming cocktail increases the yield of induced endoderm progenitors. Together, these results demonstrate the utility of our lineage-tracing method for revealing the dynamics of direct reprogramming.Combinatorial tagging of single cells using expressed DNA barcodes, delivered by a lentiviral vector, is used to track individual cells and reconstruct their lineages and trajectories during cell fate reprogramming.
中文翻译:

直接重编程中谱系和身份的单细胞图谱
直接谱系重编程涉及细胞身份的转换。单细胞技术可用于解构谱系转换过程中出现的相当大的异质性。然而,谱系关系通常会在细胞处理过程中丢失,从而使轨迹重建变得复杂。在这里,我们提出了“CellTagging”,一种组合细胞索引方法,可以并行捕获克隆历史和细胞身份,其中连续轮次的细胞标记可以构建多级谱系树。成纤维细胞诱导内胚层祖细胞重编程的细胞标记和纵向追踪揭示了两种不同的轨迹:一种导致成功重编程细胞,另一种导致“死胡同”状态,这些路径是在谱系转换的最早阶段确定的。我们发现假定的甲基转移酶 Mettl7a1 的表达与成功的重编程轨迹相关。将 Mettl7a1 添加到重编程混合物中会增加诱导内胚层祖细胞的产量。总之,这些结果证明了我们的谱系追踪方法在揭示直接重编程动态方面的实用性。使用由慢病毒载体传递的表达 DNA 条形码对单个细胞进行组合标记,用于追踪单个细胞并在重编程过程中重建其谱系和轨迹。细胞命运重编程。
更新日期:2018-12-01
中文翻译:

直接重编程中谱系和身份的单细胞图谱
直接谱系重编程涉及细胞身份的转换。单细胞技术可用于解构谱系转换过程中出现的相当大的异质性。然而,谱系关系通常会在细胞处理过程中丢失,从而使轨迹重建变得复杂。在这里,我们提出了“CellTagging”,一种组合细胞索引方法,可以并行捕获克隆历史和细胞身份,其中连续轮次的细胞标记可以构建多级谱系树。成纤维细胞诱导内胚层祖细胞重编程的细胞标记和纵向追踪揭示了两种不同的轨迹:一种导致成功重编程细胞,另一种导致“死胡同”状态,这些路径是在谱系转换的最早阶段确定的。我们发现假定的甲基转移酶 Mettl7a1 的表达与成功的重编程轨迹相关。将 Mettl7a1 添加到重编程混合物中会增加诱导内胚层祖细胞的产量。总之,这些结果证明了我们的谱系追踪方法在揭示直接重编程动态方面的实用性。使用由慢病毒载体传递的表达 DNA 条形码对单个细胞进行组合标记,用于追踪单个细胞并在重编程过程中重建其谱系和轨迹。细胞命运重编程。











































 京公网安备 11010802027423号
京公网安备 11010802027423号