当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Tuning the Performance of Synthetic Riboswitches using Machine Learning.
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2019-01-08 , DOI: 10.1021/acssynbio.8b00207 Ann-Christin Groher , Sven Jager , Christopher Schneider , Florian Groher , Kay Hamacher , Beatrix Suess
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2019-01-08 , DOI: 10.1021/acssynbio.8b00207 Ann-Christin Groher , Sven Jager , Christopher Schneider , Florian Groher , Kay Hamacher , Beatrix Suess
|
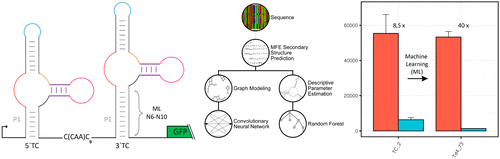
Riboswitch development for clinical, technological, and synthetic biology applications constantly seeks to optimize regulatory behavior. Here, we present a machine learning approach to improve the regulation of a tetracycline (tc)-dependent riboswitch device composed of two individual tc aptamers. We developed a bioinformatics model that combines random forest analysis with a convolutional neural network to predict the switching behavior of such tandem riboswitches. We found that both biophysical parameters and the hydrogen bond pattern influence regulation. Our new design pipeline led to significant improvement of the tc riboswitch device with a dynamic range extension from 8.5 to 40-fold. We are confident that our novel method not only results in an excellent tc-dependent riboswitch device but further holds great promise and potential for the optimization of other riboswitches.
中文翻译:

使用机器学习调整合成核糖开关的性能。
用于临床,技术和合成生物学应用的Riboswitch开发一直在寻求优化监管行为。在这里,我们提出了一种机器学习方法,以改善由两个单独的tc适体组成的依赖四环素(tc)的核糖开关设备的调节。我们开发了一种生物信息学模型,该模型结合了随机森林分析和卷积神经网络来预测此类串联核糖开关的开关行为。我们发现,生物物理参数和氢键模式都影响调节。我们的新设计流水线显着改进了tc核糖开关设备,其动态范围从8.5倍扩展到了40倍。
更新日期:2018-12-04
中文翻译:

使用机器学习调整合成核糖开关的性能。
用于临床,技术和合成生物学应用的Riboswitch开发一直在寻求优化监管行为。在这里,我们提出了一种机器学习方法,以改善由两个单独的tc适体组成的依赖四环素(tc)的核糖开关设备的调节。我们开发了一种生物信息学模型,该模型结合了随机森林分析和卷积神经网络来预测此类串联核糖开关的开关行为。我们发现,生物物理参数和氢键模式都影响调节。我们的新设计流水线显着改进了tc核糖开关设备,其动态范围从8.5倍扩展到了40倍。











































 京公网安备 11010802027423号
京公网安备 11010802027423号