当前位置:
X-MOL 学术
›
J. Phys. Chem. Lett.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Higher Accuracy Achieved in the Simulations of Protein Structure Refinement, Protein Folding, and Intrinsically Disordered Proteins Using Polarizable Force Fields
The Journal of Physical Chemistry Letters ( IF 4.8 ) Pub Date : 2018-12-05 00:00:00 , DOI: 10.1021/acs.jpclett.8b03471 Anhui Wang 1, 2 , Zhichao Zhang 2 , Guohui Li 1
The Journal of Physical Chemistry Letters ( IF 4.8 ) Pub Date : 2018-12-05 00:00:00 , DOI: 10.1021/acs.jpclett.8b03471 Anhui Wang 1, 2 , Zhichao Zhang 2 , Guohui Li 1
Affiliation
|
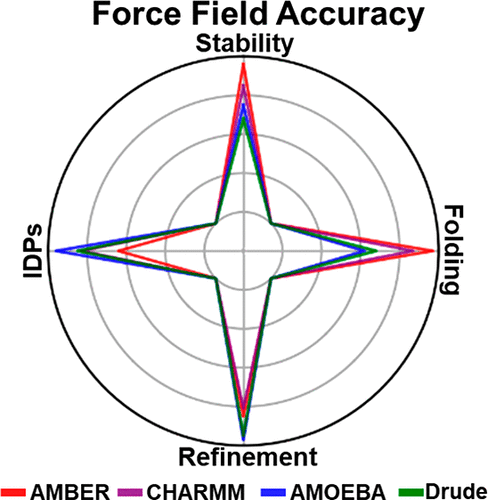
The accuracy of molecular mechanics force fields is of vital importance in biomolecular simulations. However, the admittedly more accurate polarizable force fields were recently reported to be less able to reproduce the experimental properties in comparison to additive force fields in some cases. Here, we perform long-time-scale molecular dynamics simulations to systematically evaluate the effect of explicit electronic polarization in polarizable force fields. The results show that the inclusion of electrostatic polarization effect in polarizable force fields can improve their accuracies in protein structure refinement and generate conformational ensembles more approximate to experiments for intrinsically disordered proteins. In contrast, it is difficult for polarizable force fields to approach the native structure, let alone to predict the native state when it is unknown a priori in the real protein structure predictions. We speculate that these effects might be attributed to the preference of protein–water interactions in polarizable force fields.
中文翻译:

使用极化力场在蛋白质结构细化,蛋白质折叠和固有无序蛋白质的模拟中实现了更高的精度
分子力学力场的准确性在生物分子模拟中至关重要。然而,最近报道,在某些情况下,与加成力场相比,公认的更准确的可极化力场较不具有重现实验性质的能力。在这里,我们进行长时间尺度的分子动力学模拟,以系统地评估显性电子极化在极化力场中的作用。结果表明,在极化力场中包含静电极化效应可以提高其在蛋白质结构细化中的准确性,并生成更接近于固有无序蛋白质实验的构象集合。相比之下,极化力场很难接近自然结构,真实蛋白质结构预测中的先验条件。我们推测这些影响可能归因于极化力场中蛋白质-水相互作用的偏爱。
更新日期:2018-12-05
中文翻译:

使用极化力场在蛋白质结构细化,蛋白质折叠和固有无序蛋白质的模拟中实现了更高的精度
分子力学力场的准确性在生物分子模拟中至关重要。然而,最近报道,在某些情况下,与加成力场相比,公认的更准确的可极化力场较不具有重现实验性质的能力。在这里,我们进行长时间尺度的分子动力学模拟,以系统地评估显性电子极化在极化力场中的作用。结果表明,在极化力场中包含静电极化效应可以提高其在蛋白质结构细化中的准确性,并生成更接近于固有无序蛋白质实验的构象集合。相比之下,极化力场很难接近自然结构,真实蛋白质结构预测中的先验条件。我们推测这些影响可能归因于极化力场中蛋白质-水相互作用的偏爱。











































 京公网安备 11010802027423号
京公网安备 11010802027423号