Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Kinetics of Strand Displacement and Hybridization on Wireframe DNA Nanostructures: Dissecting the Roles of Size, Morphology, and Rigidity
ACS Nano ( IF 17.1 ) Pub Date : 2018-11-28 00:00:00 , DOI: 10.1021/acsnano.8b08016 Casey M. Platnich 1 , Amani A. Hariri 1 , Janane F. Rahbani 1 , Jesse B. Gordon 1 , Hanadi F. Sleiman 1 , Gonzalo Cosa 1
ACS Nano ( IF 17.1 ) Pub Date : 2018-11-28 00:00:00 , DOI: 10.1021/acsnano.8b08016 Casey M. Platnich 1 , Amani A. Hariri 1 , Janane F. Rahbani 1 , Jesse B. Gordon 1 , Hanadi F. Sleiman 1 , Gonzalo Cosa 1
Affiliation
|
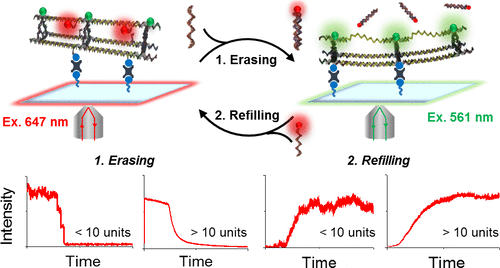
Dynamic wireframe DNA structures have gained significant attention in recent years, with research aimed toward using these architectures for sensing and encapsulation applications. For these assemblies to reach their full potential, however, knowledge of the rates of strand displacement and hybridization on these constructs is required. Herein, we report the use of single-molecule fluorescence methodologies to observe the reversible switching between double- and single-stranded forms of triangular wireframe DNA nanotubes. Specifically, by using fluorescently labeled DNA strands, we were able to monitor changes in intensity over time as we introduced different sequences. This allowed us to extract detailed kinetic information on the strand displacement and hybridization processes. Due to the polymeric nanotube structure, the ability to individually address each of the three sides, and the inherent polydispersity of our samples as a result of the step polymerization by which they are formed, a library of compounds could be studied independently yet simultaneously. Kinetic models relying on mono-exponential decays, multi-exponential decays, or sigmoidal behavior were adjusted to the different constructs to retrieve erasing and refilling kinetics. Correlations were made between the kinetic behavior observed, the site accessibility, the nanotube length, and the structural robustness of wireframe DNA nanostructures, including fully single-stranded analogs. Overall, our results reveal how the length, morphology, and rigidity of the DNA framework modulate the kinetics of strand displacement and hybridization as well as the overall addressability and structural stability of the structures under study.
中文翻译:

线框DNA纳米结构上链置换和杂交的动力学:剖析大小,形态和刚性的作用。
近年来,动态线框DNA结构受到了广泛的关注,其研究旨在将这些结构用于传感和封装应用。然而,为了使这些组装发挥最大的潜力,需要了解这些构建体上链置换和杂交的速率。在这里,我们报告使用单分子荧光方法来观察三角形线框DNA纳米管的双链和单链形式之间的可逆切换。具体来说,通过使用荧光标记的DNA链,我们能够在引入不同序列时监测强度随时间的变化。这使我们能够提取有关链置换和杂交过程的详细动力学信息。由于高分子纳米管的结构,分别解决这三个方面的能力,以及我们通过分步聚合形成样品所固有的多分散性,可以同时独立地研究化合物库。依赖于单指数衰减,多指数衰减或S形行为的动力学模型被调整为不同的构造,以恢复擦除和重新填充的动力学。在观察到的动力学行为,位点可及性,纳米管长度和线框DNA纳米结构(包括完全单链类似物)的结构坚固性之间建立了相关性。总体而言,我们的结果揭示了长度,形态,
更新日期:2018-11-28
中文翻译:

线框DNA纳米结构上链置换和杂交的动力学:剖析大小,形态和刚性的作用。
近年来,动态线框DNA结构受到了广泛的关注,其研究旨在将这些结构用于传感和封装应用。然而,为了使这些组装发挥最大的潜力,需要了解这些构建体上链置换和杂交的速率。在这里,我们报告使用单分子荧光方法来观察三角形线框DNA纳米管的双链和单链形式之间的可逆切换。具体来说,通过使用荧光标记的DNA链,我们能够在引入不同序列时监测强度随时间的变化。这使我们能够提取有关链置换和杂交过程的详细动力学信息。由于高分子纳米管的结构,分别解决这三个方面的能力,以及我们通过分步聚合形成样品所固有的多分散性,可以同时独立地研究化合物库。依赖于单指数衰减,多指数衰减或S形行为的动力学模型被调整为不同的构造,以恢复擦除和重新填充的动力学。在观察到的动力学行为,位点可及性,纳米管长度和线框DNA纳米结构(包括完全单链类似物)的结构坚固性之间建立了相关性。总体而言,我们的结果揭示了长度,形态,



























 京公网安备 11010802027423号
京公网安备 11010802027423号