Nature Methods ( IF 36.1 ) Pub Date : 2018-11-26 , DOI: 10.1038/s41592-018-0220-y Daniel Benhalevy 1 , Dimitrios G Anastasakis 1 , Markus Hafner 1
|
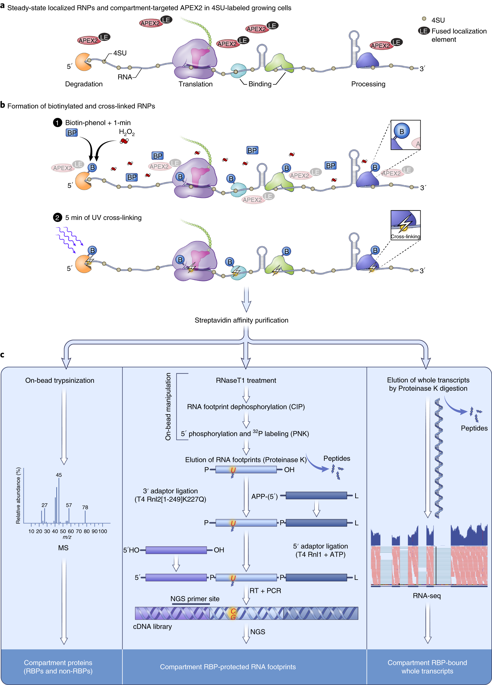
Methods for the systematic study of subcellular RNA localization are limited, and their development has lagged behind that of proteomic tools. We combined APEX2-mediated proximity biotinylation of proteins with photoactivatable ribonucleoside-enhanced crosslinking to simultaneously profile the proteome and the transcriptome bound by RNA-binding proteins in any given subcellular compartment. Our approach is fractionation independent and allows study of the localization of RNA processing intermediates, as well as the identification of regulatory RNA cis-acting elements occupied by proteins, in a cellular-compartment-specific manner. We used our method, Proximity-CLIP, to profile RNA and protein in the nucleus, in the cytoplasm, and at cell–cell interfaces. Among other insights, we observed frequent transcriptional readthrough continuing for several kilobases downstream of the canonical cleavage and polyadenylation site and a differential RBP occupancy pattern for mRNAs in the nucleus and cytoplasm. We observed that mRNAs localized to cell–cell interfaces often encoded regulatory proteins and contained protein-occupied CUG sequence elements in their 3′ untranslated region.
中文翻译:

邻近CLIP提供了亚细胞区室中蛋白质占用的RNA元素的快照。
系统地研究亚细胞RNA定位的方法是有限的,其发展落后于蛋白质组学工具。我们将APEX2介导的蛋白质的近距离生物素化与可光活化的核糖核苷增强的交联相结合,以同时分析蛋白质组和转录组,该蛋白质组和转录组由任何给定的亚细胞区室中的RNA结合蛋白结合。我们的方法是不依赖分级分离的,并允许以细胞室特异性方式研究RNA加工中间体的定位,以及鉴定蛋白质所占据的调节性RNA顺式作用元件。我们使用Proximity-CLIP方法对细胞核,细胞质以及细胞-细胞界面中的RNA和蛋白质进行了分析。在其他见解中,我们观察到频繁的转录通读持续在规范切割和聚腺苷酸化位点下游持续几千个碱基,并且在细胞核和细胞质中存在差异的RBP占用模式。我们观察到,位于细胞-细胞界面的mRNA通常编码调节蛋白,并在其3'非翻译区中包含蛋白质占用的CUG序列元素。











































 京公网安备 11010802027423号
京公网安备 11010802027423号