当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Increased Sensitivity of Mass Spectrometry by Alkaline Two-Dimensional Liquid Chromatography: Deep Cover of the Human Proteome in Gene-Centric Mode
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2018-11-19 , DOI: 10.1021/acs.jproteome.8b00754 Ekaterina V. Ilgisonis 1 , Arthur T. Kopylov 1 , Elena A. Ponomarenko 1 , Ekaterina V. Poverennaya 1 , Olga V. Tikhonova 1 , Tatiana E. Farafonova 1 , Svetlana Novikova 1 , Andrey V. Lisitsa 1 , Victor G. Zgoda 1 , Alexander I. Archakov 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2018-11-19 , DOI: 10.1021/acs.jproteome.8b00754 Ekaterina V. Ilgisonis 1 , Arthur T. Kopylov 1 , Elena A. Ponomarenko 1 , Ekaterina V. Poverennaya 1 , Olga V. Tikhonova 1 , Tatiana E. Farafonova 1 , Svetlana Novikova 1 , Andrey V. Lisitsa 1 , Victor G. Zgoda 1 , Alexander I. Archakov 1
Affiliation
|
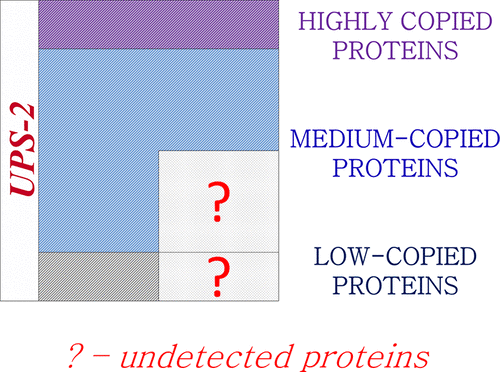
Currently, great interest is paid to the identification of “missing” proteins that have not been detected in any biological material at the protein level (PE1). In this paper, using the Universal Proteomic Standard sets 1 and 2 (UPS1 and UPS2, respectively) as an example, we characterized mass spectrometric approaches from the point of view of sensitivity (Sn), specificity (Sp), and accuracy (Ac). The aim of the paper was to show the utility of a mass spectra approach for protein detection. This sets consists of 48 high-purity human proteins without single aminoacid polymorphism (SAP) or post translational modification (PTM). The UPS1 set consists of the same 48 proteins at 5 pmols each, and in UPS2, proteins were grouped into 5 groups in accordance with their molar concentration, ranging from 10–11 to 10–6 M. Single peptides from the 92% and 96% of all sets of proteins could be detected in a pure solution of UPS2 and UPS1, respectively, by selected reaction monitoring with stable isotope-labeled standards (SRM-SIS). We also found that, in the presence of a biological matrix such as Escherichia coli extract or human blood plasma (HBP), SRM-SIS makes it possible to detect from 63% to 79% of proteins in the UPS2 set (sensitivity) with the highest specificity (∼100%) and an accuracy of 80% by increasing the sensitivity of shotgun and selected reaction monitoring combined with a stable-isotope-labeled peptide standard (SRM-SIS technology) by fractionating samples using reverse-phase liquid chromatography under alkaline conditions (2D-LC_alk). It is shown that this technique of sample fractionation allows the SRM-SIS to detect 98% of the single peptides from the proteins present in the pure solution of UPS2 (47 out of 48 proteins). When the extracts of E. coli or Pichia pastoris are added as biological matrixes to the UPS2, 46, and 45 out of 48 proteins (∼95%) can be detected, respectively, using the SRM-SIS combined with 2D-LC_alk. The combination of the 2D-LC_alk SRM-SIS and shotgun technologies allows us to increase the sensitivity up to 100% in the case of the proteins of the UPS2 set. The usage of that technology can be a solution for identifying the so-called “missing” proteins and, eventually, creating the deep proteome of a particular chromosome of tissue or organs. Experimental data have been deposited in the PeptideAtlas SRM Experiment Library with the dataset identifier PASS01192 and the PRIDE repository with the dataset identifier PXD007643.
中文翻译:

碱性二维液相色谱提高质谱的灵敏度:基因-中心模式下人类蛋白质组的深层覆盖
当前,人们非常关注鉴定在蛋白质水平(PE1)的任何生物材料中都未检测到的“缺失”蛋白质。在本文中,以通用蛋白质组学标准集1和2(分别为UPSS1和UPS2)为例,我们从灵敏度(Sn),特异性(Sp)和准确性(Ac)的角度对质谱方法进行了表征。 。本文的目的是展示质谱法在蛋白质检测中的实用性。该集合由48个高纯度人类蛋白组成,没有单个氨基酸多态性(SAP)或翻译后修饰(PTM)。UPS1集由相同的48种蛋白质组成,每种蛋白质分别为5 pmol,在UPS2中,蛋白质根据其摩尔浓度从10 –11到10分为5组。-6从92%M.单肽和蛋白质的所有组的96%可在分别UPS2和UPS1,一个纯粹的溶液通过选择的反应用稳定同位素标记的标准(SRM-SIS)监测来检测。我们还发现,在存在诸如大肠杆菌的生物基质的情况下SRM-SIS提取物或人血浆(HBP),通过增加UPSA2的特异性(〜100%)和最高80%的准确度,可以检测出63%至79%的蛋白质(灵敏度)。通过在碱性条件下(2D-LC_alk)使用反相液相色谱分离样品,将of弹枪的灵敏度和选定的反应监测与稳定的同位素标记的肽标准品(SRM-SIS技术)相结合。结果表明,这种样品分级分离技术使SRM-SIS能够从UPS2纯溶液中的蛋白质(48种蛋白质中的47种)中检测出98%的单个肽。当大肠杆菌或巴斯德毕赤酵母提取物时通过将SRM-SIS与2D-LC_alk结合使用,可将它们作为生物基质添加到UPS2中,分别可检测48种蛋白质中的46种和45种(约95%)(约95%)。结合使用2D-LC_alk SRM-SIS和shot弹枪技术,对于UPS2集的蛋白质,我们可以将灵敏度提高到100%。该技术的使用可以是一种识别所谓“缺失”蛋白质并最终创建组织或器官特定染色体的深层蛋白质组的解决方案。实验数据已保存在PeptideAtlas SRM实验库中,数据集标识符为PASS01192,而PRIDE存储库中的数据集标识符为PXD007643。
更新日期:2018-11-20
中文翻译:

碱性二维液相色谱提高质谱的灵敏度:基因-中心模式下人类蛋白质组的深层覆盖
当前,人们非常关注鉴定在蛋白质水平(PE1)的任何生物材料中都未检测到的“缺失”蛋白质。在本文中,以通用蛋白质组学标准集1和2(分别为UPSS1和UPS2)为例,我们从灵敏度(Sn),特异性(Sp)和准确性(Ac)的角度对质谱方法进行了表征。 。本文的目的是展示质谱法在蛋白质检测中的实用性。该集合由48个高纯度人类蛋白组成,没有单个氨基酸多态性(SAP)或翻译后修饰(PTM)。UPS1集由相同的48种蛋白质组成,每种蛋白质分别为5 pmol,在UPS2中,蛋白质根据其摩尔浓度从10 –11到10分为5组。-6从92%M.单肽和蛋白质的所有组的96%可在分别UPS2和UPS1,一个纯粹的溶液通过选择的反应用稳定同位素标记的标准(SRM-SIS)监测来检测。我们还发现,在存在诸如大肠杆菌的生物基质的情况下SRM-SIS提取物或人血浆(HBP),通过增加UPSA2的特异性(〜100%)和最高80%的准确度,可以检测出63%至79%的蛋白质(灵敏度)。通过在碱性条件下(2D-LC_alk)使用反相液相色谱分离样品,将of弹枪的灵敏度和选定的反应监测与稳定的同位素标记的肽标准品(SRM-SIS技术)相结合。结果表明,这种样品分级分离技术使SRM-SIS能够从UPS2纯溶液中的蛋白质(48种蛋白质中的47种)中检测出98%的单个肽。当大肠杆菌或巴斯德毕赤酵母提取物时通过将SRM-SIS与2D-LC_alk结合使用,可将它们作为生物基质添加到UPS2中,分别可检测48种蛋白质中的46种和45种(约95%)(约95%)。结合使用2D-LC_alk SRM-SIS和shot弹枪技术,对于UPS2集的蛋白质,我们可以将灵敏度提高到100%。该技术的使用可以是一种识别所谓“缺失”蛋白质并最终创建组织或器官特定染色体的深层蛋白质组的解决方案。实验数据已保存在PeptideAtlas SRM实验库中,数据集标识符为PASS01192,而PRIDE存储库中的数据集标识符为PXD007643。











































 京公网安备 11010802027423号
京公网安备 11010802027423号