当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A systematic study of minima in alanine dipeptide
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2018-10-23 , DOI: 10.1002/jcc.25589 Vladimir Mironov 1 , Yuri Alexeev 2 , Vikram Khipple Mulligan 3 , Dmitri G Fedorov 4
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2018-10-23 , DOI: 10.1002/jcc.25589 Vladimir Mironov 1 , Yuri Alexeev 2 , Vikram Khipple Mulligan 3 , Dmitri G Fedorov 4
Affiliation
|
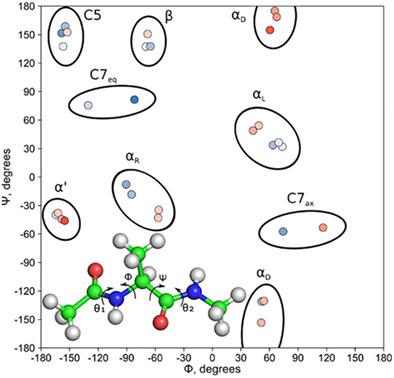
The alanine dipeptide is a standard system to model dihedral angles in proteins. It is shown that obtaining the Ramachandran plot accurately is a hard problem because of many local minima; depending on the details of geometry optimizations, different Ramachandran plots can be obtained. To locate all energy minima, starting from geometries from MD simulations, 250,000 geometry optimizations were performed at the level of RHF/6‐31G*, followed by re‐optimizations of the located 827 minima at the level of MP2/6–311++G**, yielding 30 unique minima, most of which were not previously reported in literature. Both in vacuo and solvated structures are discussed. The minima are systematically categorized based on four backbone dihedral angles. The Gibbs energies are evaluated and the structural factors determining the relative stabilities of conformers are discussed. © 2018 Wiley Periodicals, Inc.
中文翻译:

丙氨酸二肽最小值的系统研究
丙氨酸二肽是模拟蛋白质二面角的标准系统。结果表明,由于存在许多局部极小值,准确获取拉马钱德兰图是一个难题;根据几何优化的细节,可以获得不同的拉马钱德兰图。为了定位所有能量最小值,从 MD 模拟的几何形状开始,在 RHF/6-31G* 级别进行了 250,000 次几何优化,然后在 MP2/6-311++ 级别对所定位的 827 个最小值进行了重新优化G**,产生 30 个独特的最小值,其中大部分以前没有在文献中报道过。讨论了真空和溶剂化结构。基于四个骨架二面角对最小值进行系统分类。评估吉布斯能量并讨论决定构象异构体相对稳定性的结构因素。© 2018 Wiley Periodicals, Inc.
更新日期:2018-10-23
中文翻译:

丙氨酸二肽最小值的系统研究
丙氨酸二肽是模拟蛋白质二面角的标准系统。结果表明,由于存在许多局部极小值,准确获取拉马钱德兰图是一个难题;根据几何优化的细节,可以获得不同的拉马钱德兰图。为了定位所有能量最小值,从 MD 模拟的几何形状开始,在 RHF/6-31G* 级别进行了 250,000 次几何优化,然后在 MP2/6-311++ 级别对所定位的 827 个最小值进行了重新优化G**,产生 30 个独特的最小值,其中大部分以前没有在文献中报道过。讨论了真空和溶剂化结构。基于四个骨架二面角对最小值进行系统分类。评估吉布斯能量并讨论决定构象异构体相对稳定性的结构因素。© 2018 Wiley Periodicals, Inc.











































 京公网安备 11010802027423号
京公网安备 11010802027423号