Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Cryo-EM structure of the Ebola virus nucleoprotein–RNA complex at 3.6 Å resolution
Nature ( IF 50.5 ) Pub Date : 2018-10-17 , DOI: 10.1038/s41586-018-0630-0 Yukihiko Sugita 1, 2 , Hideyuki Matsunami 1 , Yoshihiro Kawaoka 3, 4, 5 , Takeshi Noda 6, 7 , Matthias Wolf 1
Nature ( IF 50.5 ) Pub Date : 2018-10-17 , DOI: 10.1038/s41586-018-0630-0 Yukihiko Sugita 1, 2 , Hideyuki Matsunami 1 , Yoshihiro Kawaoka 3, 4, 5 , Takeshi Noda 6, 7 , Matthias Wolf 1
Affiliation
|
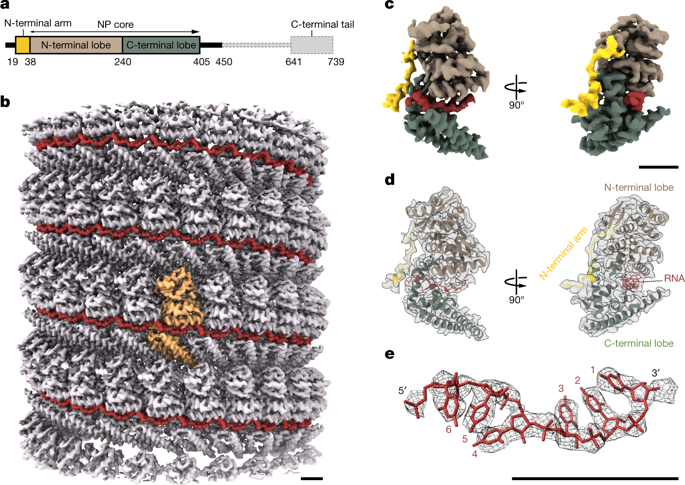
Ebola virus causes haemorrhagic fever with a high fatality rate in humans and non-human primates. It belongs to the family Filoviridae in the order Mononegavirales, which are viruses that contain linear, non-segmented, negative-sense, single-stranded genomic RNA1,2. The enveloped, filamentous virion contains the nucleocapsid, consisting of the helical nucleoprotein–RNA complex, VP24, VP30, VP35 and viral polymerase1,3. The nucleoprotein–RNA complex acts as a scaffold for nucleocapsid formation and as a template for RNA replication and transcription by condensing RNA into the virion4,5. RNA binding and nucleoprotein oligomerization are synergistic and do not readily occur independently6. Although recent cryo-electron tomography studies have revealed the overall architecture of the nucleocapsid core4,5, there has been no high-resolution reconstruction of the nucleocapsid. Here we report the structure of a recombinant Ebola virus nucleoprotein–RNA complex expressed in mammalian cells without chemical fixation, at near-atomic resolution using single-particle cryo-electron microscopy. Our structure reveals how the Ebola virus nucleocapsid core encapsidates its viral genome, its sequence-independent coordination with RNA by nucleoprotein, and the dynamic transition between the RNA-free and RNA-bound states. It provides direct structural evidence for the role of the N terminus of nucleoprotein in subunit oligomerization, and for the hydrophobic and electrostatic interactions that lead to the formation of the helical assembly. The structure is validated as representative of the native biological assembly of the nucleocapsid core by consistent dimensions and symmetry with the full virion5. The atomic model provides a detailed mechanistic basis for understanding nucleocapsid assembly and highlights key structural features that may serve as targets for anti-viral drug development.Near-atomic resolution cryo-electron microscopy structures of the Zaire ebolavirus nucleoprotein indicate a complex transition from the RNA-free to RNA-bound forms of the protein, and reveal the mechanism of oligomer formation and helical assembly.
中文翻译:

分辨率为 3.6 Å 的埃博拉病毒核蛋白-RNA 复合物的冷冻电镜结构
埃博拉病毒在人类和非人类灵长类动物中引起高致死率的出血热。它属于单线病毒目丝状病毒科,它们是含有线性、非分段、负义、单链基因组 RNA1、2 的病毒。有包膜的丝状病毒体含有核衣壳,由螺旋核蛋白-RNA 复合物、VP24、VP30、VP35 和病毒聚合酶 1、3 组成。核蛋白-RNA 复合物充当核衣壳形成的支架,并通过将 RNA 浓缩到病毒粒子中作为 RNA 复制和转录的模板 4,5。RNA 结合和核蛋白寡聚是协同的,不容易独立发生。尽管最近的低温电子断层扫描研究揭示了核衣壳核心的整体结构 4,5,没有对核衣壳进行高分辨率重建。在这里,我们报告了重组埃博拉病毒核蛋白-RNA 复合物在哺乳动物细胞中表达的结构,无需化学固定,使用单粒子冷冻电子显微镜以近原子分辨率。我们的结构揭示了埃博拉病毒核衣壳核心如何包裹其病毒基因组、核蛋白与 RNA 的序列独立协调以及无 RNA 和 RNA 结合状态之间的动态转换。它为核蛋白的 N 末端在亚基寡聚化中的作用以及导致螺旋装配体形成的疏水和静电相互作用提供了直接的结构证据。该结构通过一致的尺寸和与完整病毒粒子的对称性被验证为代表核衣壳核心的天然生物组装。原子模型为理解核衣壳组装提供了详细的机械基础,并突出了可作为抗病毒药物开发目标的关键结构特征。扎伊尔埃博拉病毒核蛋白的近原子分辨率低温电子显微镜结构表明从 RNA 的复杂转变- 游离于 RNA 结合形式的蛋白质,并揭示寡聚体形成和螺旋组装的机制。
更新日期:2018-10-17
中文翻译:

分辨率为 3.6 Å 的埃博拉病毒核蛋白-RNA 复合物的冷冻电镜结构
埃博拉病毒在人类和非人类灵长类动物中引起高致死率的出血热。它属于单线病毒目丝状病毒科,它们是含有线性、非分段、负义、单链基因组 RNA1、2 的病毒。有包膜的丝状病毒体含有核衣壳,由螺旋核蛋白-RNA 复合物、VP24、VP30、VP35 和病毒聚合酶 1、3 组成。核蛋白-RNA 复合物充当核衣壳形成的支架,并通过将 RNA 浓缩到病毒粒子中作为 RNA 复制和转录的模板 4,5。RNA 结合和核蛋白寡聚是协同的,不容易独立发生。尽管最近的低温电子断层扫描研究揭示了核衣壳核心的整体结构 4,5,没有对核衣壳进行高分辨率重建。在这里,我们报告了重组埃博拉病毒核蛋白-RNA 复合物在哺乳动物细胞中表达的结构,无需化学固定,使用单粒子冷冻电子显微镜以近原子分辨率。我们的结构揭示了埃博拉病毒核衣壳核心如何包裹其病毒基因组、核蛋白与 RNA 的序列独立协调以及无 RNA 和 RNA 结合状态之间的动态转换。它为核蛋白的 N 末端在亚基寡聚化中的作用以及导致螺旋装配体形成的疏水和静电相互作用提供了直接的结构证据。该结构通过一致的尺寸和与完整病毒粒子的对称性被验证为代表核衣壳核心的天然生物组装。原子模型为理解核衣壳组装提供了详细的机械基础,并突出了可作为抗病毒药物开发目标的关键结构特征。扎伊尔埃博拉病毒核蛋白的近原子分辨率低温电子显微镜结构表明从 RNA 的复杂转变- 游离于 RNA 结合形式的蛋白质,并揭示寡聚体形成和螺旋组装的机制。











































 京公网安备 11010802027423号
京公网安备 11010802027423号