当前位置:
X-MOL 学术
›
ACS Comb. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
PTML Combinatorial Model of ChEMBL Compounds Assays for Multiple Types of Cancer
ACS Combinatorial Science ( IF 3.903 ) Pub Date : 2018-09-21 00:00:00 , DOI: 10.1021/acscombsci.8b00090 Harbil Bediaga 1 , Sonia Arrasate 1 , Humbert González-Díaz 1, 2
ACS Combinatorial Science ( IF 3.903 ) Pub Date : 2018-09-21 00:00:00 , DOI: 10.1021/acscombsci.8b00090 Harbil Bediaga 1 , Sonia Arrasate 1 , Humbert González-Díaz 1, 2
Affiliation
|
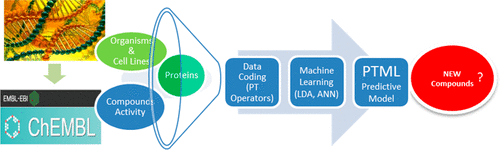
Determining the target proteins of new anticancer compounds is a very important task in Medicinal Chemistry. In this sense, chemists carry out preclinical assays with a high number of combinations of experimental conditions (cj). In fact, ChEMBL database contains outcomes of 65 534 different anticancer activity preclinical assays for 35 565 different chemical compounds (1.84 assays per compound). These assays cover different combinations of cj formed from >70 different biological activity parameters (c0), >300 different drug targets (c1), >230 cell lines (c2), and 5 organisms of assay (c3) or organisms of the target (c4). It include a total of 45 833 assays in leukemia, 6227 assays in breast cancer, 2499 assays in ovarian cancer, 3499 in colon cancer, 3159 in lung cancer, 2750 in prostate cancer, 601 in melanoma, etc. This is a very complex data set with multiple Big Data features. This data is hard to be rationalized by researchers to extract useful relationships and predict new compounds. In this context, we propose to combine perturbation theory (PT) ideas and machine learning (ML) modeling to solve this combinatorial-like problem. In this work, we report a PTML (PT + ML) model for ChEMBL data set of preclinical assays of anticancer compounds. This is a simple linear model with only three variables. The model presented values of area under receiver operating curve = AUROC = 0.872, specificity = Sp(%) = 90.2, sensitivity = Sn(%) = 70.6, and overall accuracy = Ac(%) = 87.7 in training series. The model also have Sp(%) = 90.1, Sn(%) = 71.4, and Ac(%) = 87.8 in external validation series. The model use PT operators based on multicondition moving averages to capture all the complexity of the data set. We also compared the model with nonlinear artificial neural network (ANN) models obtaining similar results. This confirms the hypothesis of a linear relationship between the PT operators and the classification as anticancer compounds in different combinations of assay conditions. Last, we compared the model with other PTML models reported in the literature concluding that this is the only one PTML model able to predict activity against multiple types of cancer. This model is a simple but versatile tool for the prediction of the targets of anticancer compounds taking into consideration multiple combinations of experimental conditions in preclinical assays.
中文翻译:

用于多种类型癌症的ChEMBL化合物测定的PTML组合模型
确定新的抗癌化合物的靶蛋白是药物化学中非常重要的任务。从这个意义上讲,化学家使用大量实验条件(c j)进行临床前测定。实际上,ChEMBL数据库包含针对35565种不同化合物的65534种不同的抗癌活性临床前测定的结果(每种化合物1.84种测定)。这些测定涵盖了由> 70种不同的生物活性参数(c 0),> 300种不同的药物靶标(c 1),> 230种细胞系(c 2)和5种测定生物(c 3)形成的c j的不同组合。)或目标生物(c 4)。它包括白血病中的45 833次检测,乳腺癌中的6227检测,卵巢癌中的2499检测,结肠癌中的3499检测,肺癌中的3159检测,前列腺癌中的2750检测,黑素瘤中的601检测等。这是一个非常复杂的数据设置了多个大数据功能。研究人员很难对这些数据进行合理化以提取有用的关系并预测新化合物。在这种情况下,我们建议将摄动理论(PT)的思想与机器学习(ML)建模相结合,以解决这种组合式问题。在这项工作中,我们报告了针对抗癌化合物临床前测定的ChEMBL数据集的PTML(PT + ML)模型。这是一个只有三个变量的简单线性模型。该模型显示了接收器工作曲线下的面积值= AUROC = 0.872,特异性= Sp(%)= 90.2,灵敏度= Sn(%)= 70.6,在训练系列中,总体准确度= Ac(%)= 87.7。在外部验证系列中,该模型还具有Sp(%)= 90.1,Sn(%)= 71.4和Ac(%)= 87.8。该模型使用基于多条件移动平均值的PT运算符来捕获数据集的所有复杂性。我们还将该模型与获得相似结果的非线性人工神经网络(ANN)模型进行了比较。这证实了在不同测定条件组合中PT操纵子与分类为抗癌化合物之间存在线性关系的假设。最后,我们将该模型与文献中报道的其他PTML模型进行了比较,得出的结论是,这是唯一一种能够预测针对多种类型癌症的活性的PTML模型。
更新日期:2018-09-21
中文翻译:

用于多种类型癌症的ChEMBL化合物测定的PTML组合模型
确定新的抗癌化合物的靶蛋白是药物化学中非常重要的任务。从这个意义上讲,化学家使用大量实验条件(c j)进行临床前测定。实际上,ChEMBL数据库包含针对35565种不同化合物的65534种不同的抗癌活性临床前测定的结果(每种化合物1.84种测定)。这些测定涵盖了由> 70种不同的生物活性参数(c 0),> 300种不同的药物靶标(c 1),> 230种细胞系(c 2)和5种测定生物(c 3)形成的c j的不同组合。)或目标生物(c 4)。它包括白血病中的45 833次检测,乳腺癌中的6227检测,卵巢癌中的2499检测,结肠癌中的3499检测,肺癌中的3159检测,前列腺癌中的2750检测,黑素瘤中的601检测等。这是一个非常复杂的数据设置了多个大数据功能。研究人员很难对这些数据进行合理化以提取有用的关系并预测新化合物。在这种情况下,我们建议将摄动理论(PT)的思想与机器学习(ML)建模相结合,以解决这种组合式问题。在这项工作中,我们报告了针对抗癌化合物临床前测定的ChEMBL数据集的PTML(PT + ML)模型。这是一个只有三个变量的简单线性模型。该模型显示了接收器工作曲线下的面积值= AUROC = 0.872,特异性= Sp(%)= 90.2,灵敏度= Sn(%)= 70.6,在训练系列中,总体准确度= Ac(%)= 87.7。在外部验证系列中,该模型还具有Sp(%)= 90.1,Sn(%)= 71.4和Ac(%)= 87.8。该模型使用基于多条件移动平均值的PT运算符来捕获数据集的所有复杂性。我们还将该模型与获得相似结果的非线性人工神经网络(ANN)模型进行了比较。这证实了在不同测定条件组合中PT操纵子与分类为抗癌化合物之间存在线性关系的假设。最后,我们将该模型与文献中报道的其他PTML模型进行了比较,得出的结论是,这是唯一一种能够预测针对多种类型癌症的活性的PTML模型。



























 京公网安备 11010802027423号
京公网安备 11010802027423号