当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Postnovo: Postprocessing Enables Accurate and FDR-Controlled de Novo Peptide Sequencing
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2018-10-16 , DOI: 10.1021/acs.jproteome.8b00278 Samuel E. Miller 1 , Adriana I. Rizzo 1 , Jacob R. Waldbauer 1
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2018-10-16 , DOI: 10.1021/acs.jproteome.8b00278 Samuel E. Miller 1 , Adriana I. Rizzo 1 , Jacob R. Waldbauer 1
Affiliation
|
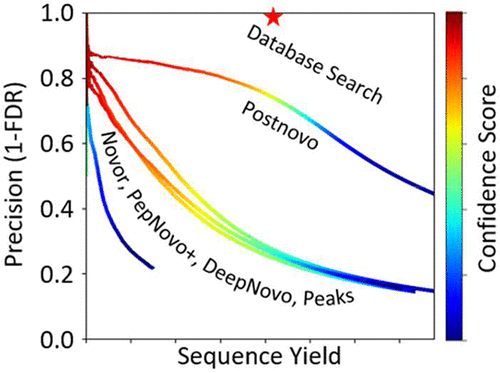
De novo sequencing offers an alternative to database search methods for peptide identification from mass spectra. Since it does not rely on a predetermined database of expected or potential sequences in the sample, de novo sequencing is particularly appropriate for samples lacking a well-defined or comprehensive reference database. However, the low accuracy of many de novo sequence predictions has prevented the widespread use of the variety of sequencing tools currently available. Here, we present a new open-source tool, Postnovo, that postprocesses de novo sequence predictions to find high-accuracy results. Postnovo uses a predictive model to rescore and rerank candidate sequences in a manner akin to database search postprocessing tools such as Percolator. Postnovo leverages the output from multiple de novo sequencing tools in its own analyses, producing many times the length of amino acid sequence information (including both full- and partial-length peptide sequences) at an equivalent false discovery rate (FDR) compared to any individual tool. We present a methodology to reliably screen the sequence predictions to a desired FDR given the Postnovo sequence score. We validate Postnovo with multiple data sets and demonstrate its ability to identify proteins that are missed by database search even in samples with paired reference databases.
中文翻译:

Postnovo:后处理可实现精确的且受FDR控制的从头测序
从头测序提供了数据库检索方法的另一种选择,可用于从质谱图中鉴定肽。因为从头测序不依赖样品中预期或潜在序列的预定数据库,所以从头测序特别适用于缺少定义明确或全面的参考数据库的样品。然而,许多从头序列预测的低准确性阻止了当前可用的各种测序工具的广泛使用。在这里,我们介绍了一个新的开源工具Postnovo,该工具对从头序列预测进行后处理,以找到高精度结果。Postnovo使用预测模型以类似于数据库搜索后处理工具(例如Percolator)的方式对候选序列进行重新评分和重新排序。Postnovo在自己的分析中利用了多个从头测序工具的输出,与任何单个工具相比,其产生的氨基酸序列信息长度(包括全长和部分全长肽序列)的长度是错误发现率(FDR)的许多倍。我们给出了一种方法,可在给定Postnovo序列评分的情况下可靠地将序列预测筛选到所需的FDR。我们使用多个数据集验证了Postnovo的性能,并证明了它具有识别数据库搜索中遗漏的蛋白质的能力,即使在具有配对参考数据库的样品中也是如此。
更新日期:2018-10-16
中文翻译:

Postnovo:后处理可实现精确的且受FDR控制的从头测序
从头测序提供了数据库检索方法的另一种选择,可用于从质谱图中鉴定肽。因为从头测序不依赖样品中预期或潜在序列的预定数据库,所以从头测序特别适用于缺少定义明确或全面的参考数据库的样品。然而,许多从头序列预测的低准确性阻止了当前可用的各种测序工具的广泛使用。在这里,我们介绍了一个新的开源工具Postnovo,该工具对从头序列预测进行后处理,以找到高精度结果。Postnovo使用预测模型以类似于数据库搜索后处理工具(例如Percolator)的方式对候选序列进行重新评分和重新排序。Postnovo在自己的分析中利用了多个从头测序工具的输出,与任何单个工具相比,其产生的氨基酸序列信息长度(包括全长和部分全长肽序列)的长度是错误发现率(FDR)的许多倍。我们给出了一种方法,可在给定Postnovo序列评分的情况下可靠地将序列预测筛选到所需的FDR。我们使用多个数据集验证了Postnovo的性能,并证明了它具有识别数据库搜索中遗漏的蛋白质的能力,即使在具有配对参考数据库的样品中也是如此。











































 京公网安备 11010802027423号
京公网安备 11010802027423号