当前位置:
X-MOL 学术
›
J. Proteome Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Integration of Transcriptomics and Antibody-Based Proteomics for Exploration of Proteins Expressed in Specialized Tissues
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2018-10-16 , DOI: 10.1021/acs.jproteome.8b00406 Evelina Sjöstedt 1, 2 , Åsa Sivertsson 1 , Feria Hikmet Noraddin 2 , Borbala Katona 2 , Åsa Näsström 2 , Jimmy Vuu 2 , Dennis Kesti 2 , Per Oksvold 1 , Per-Henrik Edqvist 2 , Ingmarie Olsson 2 , Mathias Uhlén 1 , Cecilia Lindskog 2
Journal of Proteome Research ( IF 3.8 ) Pub Date : 2018-10-16 , DOI: 10.1021/acs.jproteome.8b00406 Evelina Sjöstedt 1, 2 , Åsa Sivertsson 1 , Feria Hikmet Noraddin 2 , Borbala Katona 2 , Åsa Näsström 2 , Jimmy Vuu 2 , Dennis Kesti 2 , Per Oksvold 1 , Per-Henrik Edqvist 2 , Ingmarie Olsson 2 , Mathias Uhlén 1 , Cecilia Lindskog 2
Affiliation
|
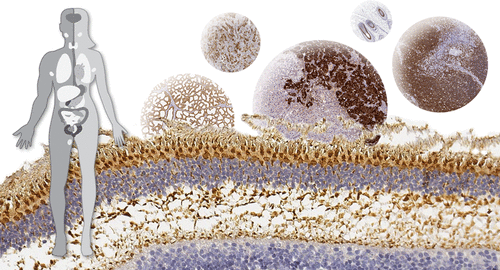
A large portion of human proteins are referred to as missing proteins, defined as protein-coding genes that lack experimental data on the protein level due to factors such as temporal expression, expression in tissues that are difficult to sample, or they actually do not encode functional proteins. In the present investigation, an integrated omics approach was used for identification and exploration of missing proteins. Transcriptomics data from three different sources—the Human Protein Atlas (HPA), the GTEx consortium, and the FANTOM5 consortium—were used as a starting point to identify genes selectively expressed in specialized tissues. Complementing the analysis with profiling on more specific tissues based on immunohistochemistry allowed for further exploration of cell-type-specific expression patterns. More detailed tissue profiling was performed for >300 genes on complementing tissues. The analysis identified tissue-specific expression of nine proteins previously listed as missing proteins (POU4F1, FRMD1, ARHGEF33, GABRG1, KRTAP2-1, BHLHE22, SPRR4, AVPR1B, and DCLK3), as well as numerous proteins with evidence of existence on the protein level that previously lacked information on spatial resolution and cell-type-specific expression pattern. We here present a comprehensive strategy for identification of missing proteins by combining transcriptomics with antibody-based proteomics. The analyzed proteins provide interesting targets for organ-specific research in health and disease.
中文翻译:

转录组学和基于抗体的蛋白质组学的整合,用于探索专业组织中表达的蛋白质
人类蛋白质的很大一部分被称为缺失蛋白质,定义为蛋白质编码基因,由于时间表达,难以在组织中表达等因素而在蛋白质水平上缺乏实验数据,或者它们实际上不编码功能蛋白。在本研究中,使用集成的组学方法来鉴定和探索缺失的蛋白质。来自三种不同来源(人类蛋白质图谱(HPA),GTEx联盟和FANTOM5联盟)的转录组学数据被用作识别在特定组织中选择性表达的基因的起点。通过对基于免疫组织化学的更特定组织进行分析来补充分析,可以进一步探索细胞类型特异性表达模式。对互补组织上的> 300个基因进行了更详细的组织分析。该分析鉴定了先前被称为缺失蛋白的9种蛋白(POU4F1,FRMD1,ARHGEF33,GABRG1,KRTAP2-1,BHLHE22,SPRR4,AVPR1B和DCLK3)的组织特异性表达,以及众多在蛋白上存在证据的蛋白以前缺乏有关空间分辨率和特定于细胞类型的表达模式的信息的水平。我们在这里提出了一种综合的策略,通过将转录组学与基于抗体的蛋白质组学相结合来鉴定缺失的蛋白质。分析的蛋白质为健康和疾病的器官特异性研究提供了有趣的靶标。FRMD1,ARHGEF33,GABRG1,KRTAP2-1,BHLHE22,SPRR4,AVPR1B和DCLK3),以及许多在蛋白质水平上存在证据的蛋白质,这些蛋白质以前缺乏有关空间分辨率和细胞类型特异性表达模式的信息。我们在这里提出了一种综合的策略,通过将转录组学与基于抗体的蛋白质组学相结合来鉴定缺失的蛋白质。分析的蛋白质为健康和疾病的器官特异性研究提供了有趣的靶标。FRMD1,ARHGEF33,GABRG1,KRTAP2-1,BHLHE22,SPRR4,AVPR1B和DCLK3),以及许多在蛋白质水平上存在证据的蛋白质,这些蛋白质以前缺乏有关空间分辨率和细胞类型特异性表达模式的信息。我们在这里提出了一种综合的策略,通过将转录组学与基于抗体的蛋白质组学相结合来鉴定缺失的蛋白质。分析的蛋白质为健康和疾病的器官特异性研究提供了有趣的靶标。我们在这里提出了一种综合的策略,通过将转录组学与基于抗体的蛋白质组学相结合来鉴定缺失的蛋白质。分析的蛋白质为健康和疾病的器官特异性研究提供了有趣的靶标。我们在这里提出了一种综合的策略,通过将转录组学与基于抗体的蛋白质组学相结合来鉴定缺失的蛋白质。分析的蛋白质为健康和疾病的器官特异性研究提供了有趣的靶标。
更新日期:2018-10-17
中文翻译:

转录组学和基于抗体的蛋白质组学的整合,用于探索专业组织中表达的蛋白质
人类蛋白质的很大一部分被称为缺失蛋白质,定义为蛋白质编码基因,由于时间表达,难以在组织中表达等因素而在蛋白质水平上缺乏实验数据,或者它们实际上不编码功能蛋白。在本研究中,使用集成的组学方法来鉴定和探索缺失的蛋白质。来自三种不同来源(人类蛋白质图谱(HPA),GTEx联盟和FANTOM5联盟)的转录组学数据被用作识别在特定组织中选择性表达的基因的起点。通过对基于免疫组织化学的更特定组织进行分析来补充分析,可以进一步探索细胞类型特异性表达模式。对互补组织上的> 300个基因进行了更详细的组织分析。该分析鉴定了先前被称为缺失蛋白的9种蛋白(POU4F1,FRMD1,ARHGEF33,GABRG1,KRTAP2-1,BHLHE22,SPRR4,AVPR1B和DCLK3)的组织特异性表达,以及众多在蛋白上存在证据的蛋白以前缺乏有关空间分辨率和特定于细胞类型的表达模式的信息的水平。我们在这里提出了一种综合的策略,通过将转录组学与基于抗体的蛋白质组学相结合来鉴定缺失的蛋白质。分析的蛋白质为健康和疾病的器官特异性研究提供了有趣的靶标。FRMD1,ARHGEF33,GABRG1,KRTAP2-1,BHLHE22,SPRR4,AVPR1B和DCLK3),以及许多在蛋白质水平上存在证据的蛋白质,这些蛋白质以前缺乏有关空间分辨率和细胞类型特异性表达模式的信息。我们在这里提出了一种综合的策略,通过将转录组学与基于抗体的蛋白质组学相结合来鉴定缺失的蛋白质。分析的蛋白质为健康和疾病的器官特异性研究提供了有趣的靶标。FRMD1,ARHGEF33,GABRG1,KRTAP2-1,BHLHE22,SPRR4,AVPR1B和DCLK3),以及许多在蛋白质水平上存在证据的蛋白质,这些蛋白质以前缺乏有关空间分辨率和细胞类型特异性表达模式的信息。我们在这里提出了一种综合的策略,通过将转录组学与基于抗体的蛋白质组学相结合来鉴定缺失的蛋白质。分析的蛋白质为健康和疾病的器官特异性研究提供了有趣的靶标。我们在这里提出了一种综合的策略,通过将转录组学与基于抗体的蛋白质组学相结合来鉴定缺失的蛋白质。分析的蛋白质为健康和疾病的器官特异性研究提供了有趣的靶标。我们在这里提出了一种综合的策略,通过将转录组学与基于抗体的蛋白质组学相结合来鉴定缺失的蛋白质。分析的蛋白质为健康和疾病的器官特异性研究提供了有趣的靶标。











































 京公网安备 11010802027423号
京公网安备 11010802027423号