Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
On-Axis Alignment of Protein Nanocage Assemblies from 2D to 3D through the Aromatic Stacking Interactions of Amino Acid Residues
ACS Nano ( IF 15.8 ) Pub Date : 2018-09-28 00:00:00 , DOI: 10.1021/acsnano.8b06091 Kai Zhou 1 , Jiachen Zang 1 , Hai Chen 1 , Wenming Wang 2 , Hongfei Wang 2 , Guanghua Zhao 1
ACS Nano ( IF 15.8 ) Pub Date : 2018-09-28 00:00:00 , DOI: 10.1021/acsnano.8b06091 Kai Zhou 1 , Jiachen Zang 1 , Hai Chen 1 , Wenming Wang 2 , Hongfei Wang 2 , Guanghua Zhao 1
Affiliation
|
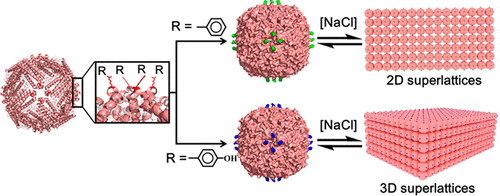
Aromatic–aromatic interactions between natural aromatic amino acids Phe, Tyr, and Trp play crucial roles in protein–protein recognition and protein folding. However, the function of such interactions in the preparation of different dimensional, ordered protein superstructures has not been recognized. Herein, by a combination of the directionality of the symmetry axes of protein building blocks and the strength of the aromatic–aromatic interactions coming from a group of aromatic amino acid residues, we built an engineering strategy to construct protein superlattices. Based on this strategy, substitution of single amino acid residue Glu162 around the C4 rotation axes near the outer surface of 24-mer ferritin nanocage with Phe, Tyr, and Trp, respectively, resulted in 2D and 3D protein superlattices where protein cages are aligned along the C4 axes, imposing a fixed disposition of neighboring ferritins. The self-assembly of these superlattices is reversible, which can be tuned by external stimuli (salt concentration or pH). Moreover, these superlattices can serve as biotemplates for the fabrication of 2D and 3D inorganic nanoparticle arrays.
中文翻译:

通过氨基酸残基的芳香堆积相互作用将蛋白质纳米笼组件从2D对齐到3D。
天然芳香氨基酸Phe,Tyr和Trp之间的芳香-芳香相互作用在蛋白质-蛋白质识别和蛋白质折叠中起关键作用。然而,这种相互作用在制备不同尺寸的,有序的蛋白质超结构中的功能尚未得到认识。在这里,结合蛋白质构件的对称轴的方向性和来自一组芳香族氨基酸残基的芳香族-芳香族相互作用的强度,我们构建了一种构建蛋白质超晶格的工程策略。基于此策略,在C 4周围取代单个氨基酸残基Glu162分别与Phe,Tyr和Trp结合的24-mer铁蛋白纳米笼的外表面附近的旋转轴导致2D和3D蛋白质超晶格,其中蛋白笼沿C 4轴对齐,从而对邻近的铁蛋白进行固定定位。这些超晶格的自组装是可逆的,可以通过外部刺激(盐浓度或pH)来调节。而且,这些超晶格可以用作制造2D和3D无机纳米粒子阵列的生物模板。
更新日期:2018-09-28
中文翻译:

通过氨基酸残基的芳香堆积相互作用将蛋白质纳米笼组件从2D对齐到3D。
天然芳香氨基酸Phe,Tyr和Trp之间的芳香-芳香相互作用在蛋白质-蛋白质识别和蛋白质折叠中起关键作用。然而,这种相互作用在制备不同尺寸的,有序的蛋白质超结构中的功能尚未得到认识。在这里,结合蛋白质构件的对称轴的方向性和来自一组芳香族氨基酸残基的芳香族-芳香族相互作用的强度,我们构建了一种构建蛋白质超晶格的工程策略。基于此策略,在C 4周围取代单个氨基酸残基Glu162分别与Phe,Tyr和Trp结合的24-mer铁蛋白纳米笼的外表面附近的旋转轴导致2D和3D蛋白质超晶格,其中蛋白笼沿C 4轴对齐,从而对邻近的铁蛋白进行固定定位。这些超晶格的自组装是可逆的,可以通过外部刺激(盐浓度或pH)来调节。而且,这些超晶格可以用作制造2D和3D无机纳米粒子阵列的生物模板。











































 京公网安备 11010802027423号
京公网安备 11010802027423号