当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Retroelement-Based Genome Editing and Evolution
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-09-26 00:00:00 , DOI: 10.1021/acssynbio.8b00273 Anna J. Simon , Barrett R. Morrow , Andrew D. Ellington
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-09-26 00:00:00 , DOI: 10.1021/acssynbio.8b00273 Anna J. Simon , Barrett R. Morrow , Andrew D. Ellington
|
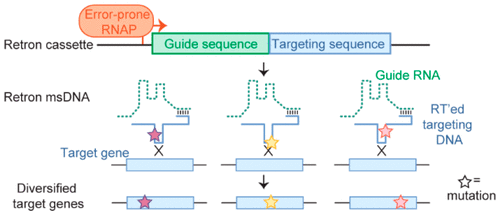
While several genome editing methods exist, few are suitable for the continuous evolution of targeted sequences. Here we develop bacterial retroelements known as “retrons” for the dynamic, in vivo editing and mutagenesis of targeted genes. We first optimized retrons’ ability to introduce preprogrammed mutations, optimizing both their expression and the host machinery that interacts with them to increase the incorporation frequency of mutations 78-fold over rates previously reported in synthetic systems. The optimized system is capable of simultaneously overwriting 13 separate positions spanning a 31-base length, and is for the first time shown to yield targeted deletions and insertions. To engineer retrons as a tool to introduce novel, unprogrammed mutations in specific targeted regions, we expressed them under a mutagenic T7 RNA polymerase. This coupled mutagenic T7 RNA polymerase-retron system enabled the evolution of diverse variants of environmentally selected antibiotic resistance genes, producing mutation rates in the targeted region 190-fold higher than background cellular mutation rates, potentially enabling the dynamic, continuous self-evolution of selected phenotypes.
中文翻译:

基于Retroelement的基因组编辑和进化
虽然存在几种基因组编辑方法,但很少有方法适合连续进化目标序列。在这里,我们为体内的动态细菌开发了被称为“ retrons”的细菌改造元件 。靶向基因的编辑和诱变。我们首先优化了Retrons引入预编程突变的能力,同时优化了它们的表达和与它们相互作用的宿主机制,从而使突变的掺入频率比以前在合成系统中报道的速率增加了78倍。优化的系统能够同时覆盖13个碱基长度的13个独立位置,并且首次显示出靶向的缺失和插入。为了工程改造作为在特定目标区域引入新的,未编程的突变的工具,我们在诱变的T7 RNA聚合酶下表达了它们。这种耦合的诱变T7 RNA聚合酶-还原剂系统使环境选择的抗生素抗性基因的多种变体得以进化,
更新日期:2018-09-26
中文翻译:

基于Retroelement的基因组编辑和进化
虽然存在几种基因组编辑方法,但很少有方法适合连续进化目标序列。在这里,我们为体内的动态细菌开发了被称为“ retrons”的细菌改造元件 。靶向基因的编辑和诱变。我们首先优化了Retrons引入预编程突变的能力,同时优化了它们的表达和与它们相互作用的宿主机制,从而使突变的掺入频率比以前在合成系统中报道的速率增加了78倍。优化的系统能够同时覆盖13个碱基长度的13个独立位置,并且首次显示出靶向的缺失和插入。为了工程改造作为在特定目标区域引入新的,未编程的突变的工具,我们在诱变的T7 RNA聚合酶下表达了它们。这种耦合的诱变T7 RNA聚合酶-还原剂系统使环境选择的抗生素抗性基因的多种变体得以进化,











































 京公网安备 11010802027423号
京公网安备 11010802027423号