Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Principles of nucleosome organization revealed by single-cell micrococcal nuclease sequencing
Nature ( IF 50.5 ) Pub Date : 2018-09-26 , DOI: 10.1038/s41586-018-0567-3 Binbin Lai 1 , Weiwu Gao 1, 2 , Kairong Cui 1 , Wanli Xie 1, 3 , Qingsong Tang 1 , Wenfei Jin 4 , Gangqing Hu 1 , Bing Ni 2 , Keji Zhao 1
Nature ( IF 50.5 ) Pub Date : 2018-09-26 , DOI: 10.1038/s41586-018-0567-3 Binbin Lai 1 , Weiwu Gao 1, 2 , Kairong Cui 1 , Wanli Xie 1, 3 , Qingsong Tang 1 , Wenfei Jin 4 , Gangqing Hu 1 , Bing Ni 2 , Keji Zhao 1
Affiliation
|
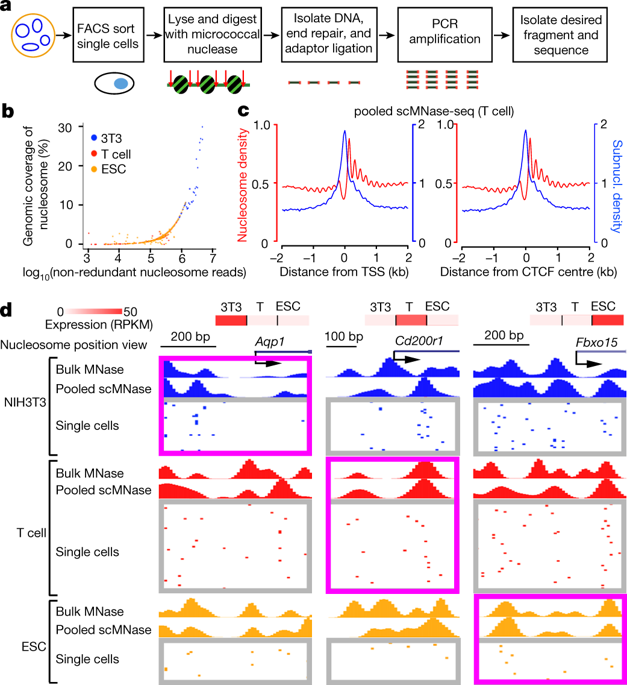
Nucleosome positioning is critical to chromatin accessibility and is associated with gene expression programs in cells1–3. Previous nucleosome mapping methods assemble profiles from cell populations and reveal a cell-averaged pattern: nucleosomes are positioned and form a phased array that surrounds the transcription start sites of active genes3–6 and DNase I hypersensitive sites7. However, even in a homogenous population of cells, cells exhibit heterogeneity in expression in response to active signalling8,9 that may be related to heterogeneity in chromatin accessibility10–12. Here we report a technique, termed single-cell micrococcal nuclease sequencing (scMNase-seq), that can be used to simultaneously measure genome-wide nucleosome positioning and chromatin accessibility in single cells. Application of scMNase-seq to NIH3T3 cells, mouse primary naive CD4 T cells and mouse embryonic stem cells reveals two principles of nucleosome organization: first, nucleosomes in heterochromatin regions, or that surround the transcription start sites of silent genes, show large variation in positioning across different cells but are highly uniformly spaced along the nucleosome array; and second, nucleosomes that surround the transcription start sites of active genes and DNase I hypersensitive sites show little variation in positioning across different cells but are relatively heterogeneously spaced along the nucleosome array. We found a bimodal distribution of nucleosome spacing at DNase I hypersensitive sites, which corresponds to inaccessible and accessible states and is associated with nucleosome variation and variation in accessibility across cells. Nucleosome variation is smaller within single cells than across cells, and smaller within the same cell type than across cell types. A large fraction of naive CD4 T cells and mouse embryonic stem cells shows depleted nucleosome occupancy at the de novo enhancers detected in their respective differentiated lineages, revealing the existence of cells primed for differentiation to specific lineages in undifferentiated cell populations.Single-cell micrococcal nuclease sequencing simultaneously measures chromatin accessibility and genome-wide nucleosome positioning in single cells to reveal principles of nucleosome organization.
中文翻译:

单细胞微球菌核酸酶测序揭示的核小体组织原理
核小体定位对于染色质可及性至关重要,并且与细胞 1-3 中的基因表达程序相关。以前的核小体作图方法从细胞群中组装图谱并揭示细胞平均模式:核小体被定位并形成一个相控阵,围绕着活性基因 3-6 的转录起始位点和 DNase I 超敏位点 7。然而,即使在同质的细胞群中,细胞也表现出响应活动信号的表达异质性 8,9,这可能与染色质可及性的异质性 10-12 相关。在这里,我们报告了一种称为单细胞微球菌核酸酶测序 (scMNase-seq) 的技术,该技术可用于同时测量单细胞中的全基因组核小体定位和染色质可及性。scMNase-seq在NIH3T3细胞中的应用,小鼠原代 CD4 T 细胞和小鼠胚胎干细胞揭示了核小体组织的两个原则:首先,异染色质区域中的核小体,或围绕沉默基因转录起始位点的核小体,在不同细胞间的定位差异很大,但沿核小体阵列;其次,围绕活性基因转录起始位点和 DNase I 超敏位点的核小体在不同细胞中的定位几乎没有变化,但沿核小体阵列的间隔相对不均匀。我们在 DNase I 超敏感位点发现核小体间距的双峰分布,这对应于不可接近和可接近的状态,并且与核小体变异和跨细胞可接近性的变异有关。单细胞内的核小体变异小于细胞间的变异,同一细胞类型内的核小体变异小于细胞类型间的变异。大部分幼稚 CD4 T 细胞和小鼠胚胎干细胞在其各自分化谱系中检测到的 de novo 增强子处显示耗尽的核小体占据,揭示了在未分化细胞群中存在准备分化为特定谱系的细胞。单细胞微球菌核酸酶测序同时测量单细胞中的染色质可及性和全基因组核小体定位,以揭示核小体组织的原理。
更新日期:2018-09-26
中文翻译:

单细胞微球菌核酸酶测序揭示的核小体组织原理
核小体定位对于染色质可及性至关重要,并且与细胞 1-3 中的基因表达程序相关。以前的核小体作图方法从细胞群中组装图谱并揭示细胞平均模式:核小体被定位并形成一个相控阵,围绕着活性基因 3-6 的转录起始位点和 DNase I 超敏位点 7。然而,即使在同质的细胞群中,细胞也表现出响应活动信号的表达异质性 8,9,这可能与染色质可及性的异质性 10-12 相关。在这里,我们报告了一种称为单细胞微球菌核酸酶测序 (scMNase-seq) 的技术,该技术可用于同时测量单细胞中的全基因组核小体定位和染色质可及性。scMNase-seq在NIH3T3细胞中的应用,小鼠原代 CD4 T 细胞和小鼠胚胎干细胞揭示了核小体组织的两个原则:首先,异染色质区域中的核小体,或围绕沉默基因转录起始位点的核小体,在不同细胞间的定位差异很大,但沿核小体阵列;其次,围绕活性基因转录起始位点和 DNase I 超敏位点的核小体在不同细胞中的定位几乎没有变化,但沿核小体阵列的间隔相对不均匀。我们在 DNase I 超敏感位点发现核小体间距的双峰分布,这对应于不可接近和可接近的状态,并且与核小体变异和跨细胞可接近性的变异有关。单细胞内的核小体变异小于细胞间的变异,同一细胞类型内的核小体变异小于细胞类型间的变异。大部分幼稚 CD4 T 细胞和小鼠胚胎干细胞在其各自分化谱系中检测到的 de novo 增强子处显示耗尽的核小体占据,揭示了在未分化细胞群中存在准备分化为特定谱系的细胞。单细胞微球菌核酸酶测序同时测量单细胞中的染色质可及性和全基因组核小体定位,以揭示核小体组织的原理。











































 京公网安备 11010802027423号
京公网安备 11010802027423号