当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Template-Guided Protein Structure Prediction and Refinement Using Optimized Folding Landscape Force Fields
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2018-09-21 00:00:00 , DOI: 10.1021/acs.jctc.8b00683 Mingchen Chen 1, 2 , Xingcheng Lin 1, 3 , Wei Lu 1, 3 , Nicholas P. Schafer 1, 4 , José N. Onuchic 1, 3, 4, 5 , Peter G. Wolynes 1, 4, 5
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2018-09-21 00:00:00 , DOI: 10.1021/acs.jctc.8b00683 Mingchen Chen 1, 2 , Xingcheng Lin 1, 3 , Wei Lu 1, 3 , Nicholas P. Schafer 1, 4 , José N. Onuchic 1, 3, 4, 5 , Peter G. Wolynes 1, 4, 5
Affiliation
|
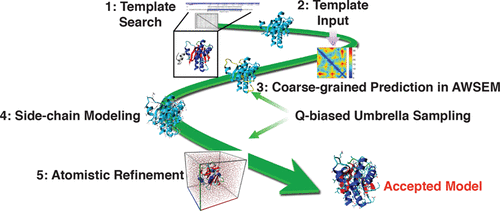
When good structural templates can be identified, template-based modeling is the most reliable way to predict the tertiary structure of proteins. In this study, we combine template-based modeling with a realistic coarse-grained force field, AWSEM, that has been optimized using the principles of energy landscape theory. The Associative memory, Water mediated, Structure and Energy Model (AWSEM) is a coarse-grained force field having both transferable tertiary interactions and knowledge-based local-in-sequence interaction terms. We incorporate template information into AWSEM by introducing soft collective biases to the template structures, resulting in a model that we call AWSEM-Template. Structure prediction tests on eight targets, four of which are in the low sequence identity “twilight zone” of homology modeling, show that AWSEM-Template can achieve high-resolution structure prediction. Our results also confirm that using a combination of AWSEM and a template-guided potential leads to more accurate prediction of protein structures than simply using a template-guided potential alone. Free energy profile analyses demonstrate that the soft collective biases to the template effectively increase funneling toward native-like structures while still allowing significant flexibility so as to allow for correction of discrepancies between the target structure and the template. A further stage of refinement using all-atom molecular dynamics augmented with soft collective biases to the structures predicted by AWSEM-Template leads to a further improvement of both backbone and side-chain accuracy by maintaining sufficient flexibility but at the same time discouraging unproductive unfolding events often seen in unrestrained all-atom refinement simulations. The all-atom refinement simulations also reduce patches of frustration of the initial predictions. Some of the backbones found among the structures produced during the initial coarse-grained prediction step already have CE-RMSD values of less than 3 Å with 90% or more of the residues aligned to the experimentally solved structure for all targets. All-atom structures generated during the following all-atom refinement simulations, which started from coarse-grained structures that were chosen without reference to any knowledge about the native structure, have CE-RMSD values of less than 2.5 Å with 90% or more of the residues aligned for 6 out of 8 targets. Clustering low energy structures generated during the initial coarse-grained annealing picks out reliably structures that are within 1 Å of the best sampled structures in 5 out of 8 cases. After the all-atom refinement, structures that are within 1 Å of the best sampled structures can be selected using a simple algorithm based on energetic features alone in 7 out of 8 cases.
中文翻译:

模板指导的蛋白质结构预测和优化使用优化的折叠景观力场。
当可以确定好的结构模板时,基于模板的建模是预测蛋白质三级结构的最可靠方法。在这项研究中,我们将基于模板的建模与现实的粗粒度力场AWSEM结合起来,该力场已使用能源景观理论的原理进行了优化。联想记忆,水介导的结构和能量模型(AWSEM)是一种粗粒度的力场,具有可传输的三次相互作用和基于知识的按顺序进行的局部相互作用。我们通过向模板结构引入软的集体偏见将模板信息整合到AWSEM中,从而形成一个称为AWSEM-Template的模型。对八个目标进行结构预测测试,其中四个处于同源性建模的低序列同一性“暮光区”,表明AWSEM-Template可以实现高分辨率的结构预测。我们的结果还证实,与仅使用模板指导电势相比,结合使用AWSEM和模板指导电势可以更准确地预测蛋白质结构。自由能曲线分析表明,对模板的软集合偏差有效地增加了向天然结构的漏斗,同时仍然允许显着的灵活性,从而可以校正目标结构和模板之间的差异。使用全原子分子动力学进一步完善的阶段,通过对AWSEM-Template预测的结构施加软的集体偏见来增强,从而通过保持足够的灵活性进一步抑制主链和侧链的准确性,同时又阻止了无用的展开事件通常在无限制的全原子细化模拟中看到。全原子细化模拟还减少了初始预测的挫败感。在最初的粗粒度预测步骤中生成的结构中发现的某些主干已经具有小于3的CE-RMSD值,其中90%或更多的残基与所有目标的实验解析结构对齐。在以下所有原子精炼模拟中生成的所有原子结构,从粗粒度结构开始的,这些粗粒度结构是在不参考任何天然结构知识的情况下选择的,其CE-RMSD值小于2.5Å,其中90%或更多的残基针对8个靶中的6个进行了比对。在最初的粗粒度退火过程中生成的聚集低能结构可以在8个案例中的5个案例中,从最佳采样结构的1Å范围内挑选出可靠的结构。在对所有原子进行细化之后,可以使用简单的算法,仅基于高能特征,在8种情况中的7种情况下,选择最佳采样结构的1Å以内的结构。在最初的粗粒度退火过程中生成的聚集低能结构可以在8个案例中的5个案例中,从最佳采样结构的1Å范围内挑选出可靠的结构。在对所有原子进行细化之后,可以使用简单的算法,仅基于高能特征,在8种情况中的7种情况下,选择最佳采样结构的1Å以内的结构。在最初的粗粒度退火过程中生成的聚集低能结构可以在8个案例中的5个案例中,从最佳采样结构的1Å范围内挑选出可靠的结构。在对所有原子进行细化之后,可以使用简单的算法,仅基于高能特征,在8种情况中的7种情况下,选择最佳采样结构的1Å以内的结构。
更新日期:2018-09-21
中文翻译:

模板指导的蛋白质结构预测和优化使用优化的折叠景观力场。
当可以确定好的结构模板时,基于模板的建模是预测蛋白质三级结构的最可靠方法。在这项研究中,我们将基于模板的建模与现实的粗粒度力场AWSEM结合起来,该力场已使用能源景观理论的原理进行了优化。联想记忆,水介导的结构和能量模型(AWSEM)是一种粗粒度的力场,具有可传输的三次相互作用和基于知识的按顺序进行的局部相互作用。我们通过向模板结构引入软的集体偏见将模板信息整合到AWSEM中,从而形成一个称为AWSEM-Template的模型。对八个目标进行结构预测测试,其中四个处于同源性建模的低序列同一性“暮光区”,表明AWSEM-Template可以实现高分辨率的结构预测。我们的结果还证实,与仅使用模板指导电势相比,结合使用AWSEM和模板指导电势可以更准确地预测蛋白质结构。自由能曲线分析表明,对模板的软集合偏差有效地增加了向天然结构的漏斗,同时仍然允许显着的灵活性,从而可以校正目标结构和模板之间的差异。使用全原子分子动力学进一步完善的阶段,通过对AWSEM-Template预测的结构施加软的集体偏见来增强,从而通过保持足够的灵活性进一步抑制主链和侧链的准确性,同时又阻止了无用的展开事件通常在无限制的全原子细化模拟中看到。全原子细化模拟还减少了初始预测的挫败感。在最初的粗粒度预测步骤中生成的结构中发现的某些主干已经具有小于3的CE-RMSD值,其中90%或更多的残基与所有目标的实验解析结构对齐。在以下所有原子精炼模拟中生成的所有原子结构,从粗粒度结构开始的,这些粗粒度结构是在不参考任何天然结构知识的情况下选择的,其CE-RMSD值小于2.5Å,其中90%或更多的残基针对8个靶中的6个进行了比对。在最初的粗粒度退火过程中生成的聚集低能结构可以在8个案例中的5个案例中,从最佳采样结构的1Å范围内挑选出可靠的结构。在对所有原子进行细化之后,可以使用简单的算法,仅基于高能特征,在8种情况中的7种情况下,选择最佳采样结构的1Å以内的结构。在最初的粗粒度退火过程中生成的聚集低能结构可以在8个案例中的5个案例中,从最佳采样结构的1Å范围内挑选出可靠的结构。在对所有原子进行细化之后,可以使用简单的算法,仅基于高能特征,在8种情况中的7种情况下,选择最佳采样结构的1Å以内的结构。在最初的粗粒度退火过程中生成的聚集低能结构可以在8个案例中的5个案例中,从最佳采样结构的1Å范围内挑选出可靠的结构。在对所有原子进行细化之后,可以使用简单的算法,仅基于高能特征,在8种情况中的7种情况下,选择最佳采样结构的1Å以内的结构。











































 京公网安备 11010802027423号
京公网安备 11010802027423号