当前位置:
X-MOL 学术
›
Nano Lett.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
High-Precision Single-Molecule Characterization of the Folding of an HIV RNA Hairpin by Atomic Force Microscopy
Nano Letters ( IF 9.6 ) Pub Date : 2018-09-20 00:00:00 , DOI: 10.1021/acs.nanolett.8b02597 Robert Walder 1 , William J. Van Patten 1 , Dustin B. Ritchie 2 , Rebecca K. Montange 1 , Ty W. Miller 1 , Michael T. Woodside 2 , Thomas T. Perkins 1, 3
Nano Letters ( IF 9.6 ) Pub Date : 2018-09-20 00:00:00 , DOI: 10.1021/acs.nanolett.8b02597 Robert Walder 1 , William J. Van Patten 1 , Dustin B. Ritchie 2 , Rebecca K. Montange 1 , Ty W. Miller 1 , Michael T. Woodside 2 , Thomas T. Perkins 1, 3
Affiliation
|
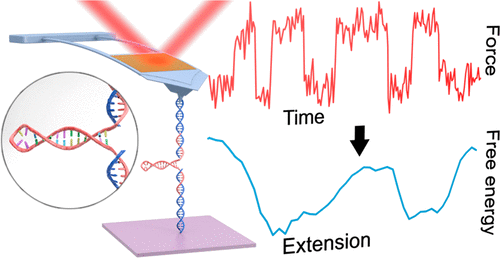
The folding of RNA into a wide range of structures is essential for its diverse biological functions from enzymatic catalysis to ligand binding and gene regulation. The unfolding and refolding of individual RNA molecules can be probed by single-molecule force spectroscopy (SMFS), enabling detailed characterization of the conformational dynamics of the molecule as well as the free-energy landscape underlying folding. Historically, high-precision SMFS studies of RNA have been limited to custom-built optical traps. Although commercial atomic force microscopes (AFMs) are widely deployed and offer significant advantages in ease-of-use over custom-built optical traps, traditional AFM-based SMFS lacks the sensitivity and stability to characterize individual RNA molecules precisely. Here, we developed a high-precision SMFS assay to study RNA folding using a commercial AFM and applied it to characterize a small RNA hairpin from HIV that plays a key role in stimulating programmed ribosomal frameshifting. We achieved rapid data acquisition in a dynamic assay, unfolding and then refolding the same individual hairpin more than 1,100 times in 15 min. In comparison to measurements using optical traps, our AFM-based assay featured a stiffer force probe and a less compliant construct, providing a complementary measurement regime that dramatically accelerated equilibrium folding dynamics. Not only did kinetic analysis of equilibrium trajectories of the HIV RNA hairpin yield the traditional parameters used to characterize folding by SMFS (zero-force rate constants and distances to the transition state), but we also reconstructed the full 1D projection of the folding free-energy landscape comparable to state-of-the-art studies using dual-beam optical traps, a first for this RNA hairpin and AFM studies of nucleic acids in general. Looking forward, we anticipate that the ease-of-use of our high-precision assay implemented on a commercial AFM will accelerate studying folding of diverse nucleic acid structures.
中文翻译:

通过原子力显微镜对HIV RNA发夹折叠的高精度单分子表征
从酶催化到配体结合和基因调控,RNA折叠成各种结构对于其多种生物学功能至关重要。单个RNA分子的解折叠和重折叠可以通过单分子力谱(SMFS)进行探测,从而能够详细表征分子的构象动力学以及折叠背后的自由能态势。从历史上看,RNA的高精度SMFS研究仅限于定制的光阱。尽管商业原子力显微镜(AFM)已被广泛部署,并且在易用性方面优于定制的光学陷阱,但传统的基于AFM的SMFS缺乏精确表征单个RNA分子的灵敏度和稳定性。这里,我们开发了一种高精度SMFS测定法,以使用商业AFM研究RNA折叠,并将其用于表征HIV的小RNA发夹,该发夹在刺激程序性核糖体移码中起关键作用。我们在动态测定中实现了快速的数据采集,将同一单个发夹展开,然后在15分钟内折叠了1100次以上。与使用光阱的测量相比,我们基于原子力显微镜的分析具有更强力的探针和不那么顺应的结构,可提供互补的测量方式,从而显着加速平衡折叠动力学。HIV RNA发夹的平衡轨迹的动力学分析不仅产生了用于表征SMFS折叠的传统参数(零力速率常数和到过渡态的距离),但我们也使用双光束光阱重建了可折叠自由能景观的完整一维投影,这与最新研究相当,这是该RNA发夹和一般AFM核酸研究的首次。展望未来,我们预计在商业AFM上实现的高精度测定的易用性将加速研究各种核酸结构的折叠。
更新日期:2018-09-20
中文翻译:

通过原子力显微镜对HIV RNA发夹折叠的高精度单分子表征
从酶催化到配体结合和基因调控,RNA折叠成各种结构对于其多种生物学功能至关重要。单个RNA分子的解折叠和重折叠可以通过单分子力谱(SMFS)进行探测,从而能够详细表征分子的构象动力学以及折叠背后的自由能态势。从历史上看,RNA的高精度SMFS研究仅限于定制的光阱。尽管商业原子力显微镜(AFM)已被广泛部署,并且在易用性方面优于定制的光学陷阱,但传统的基于AFM的SMFS缺乏精确表征单个RNA分子的灵敏度和稳定性。这里,我们开发了一种高精度SMFS测定法,以使用商业AFM研究RNA折叠,并将其用于表征HIV的小RNA发夹,该发夹在刺激程序性核糖体移码中起关键作用。我们在动态测定中实现了快速的数据采集,将同一单个发夹展开,然后在15分钟内折叠了1100次以上。与使用光阱的测量相比,我们基于原子力显微镜的分析具有更强力的探针和不那么顺应的结构,可提供互补的测量方式,从而显着加速平衡折叠动力学。HIV RNA发夹的平衡轨迹的动力学分析不仅产生了用于表征SMFS折叠的传统参数(零力速率常数和到过渡态的距离),但我们也使用双光束光阱重建了可折叠自由能景观的完整一维投影,这与最新研究相当,这是该RNA发夹和一般AFM核酸研究的首次。展望未来,我们预计在商业AFM上实现的高精度测定的易用性将加速研究各种核酸结构的折叠。











































 京公网安备 11010802027423号
京公网安备 11010802027423号