当前位置:
X-MOL 学术
›
J. Chem. Educ.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Program for Simulating Gel Electrophoresis of Enzyme-Digested Proteins
Journal of Chemical Education ( IF 2.5 ) Pub Date : 2018-09-14 00:00:00 , DOI: 10.1021/acs.jchemed.8b00225 Howard Mayes 1 , Chung F. Wong 1, 2
Journal of Chemical Education ( IF 2.5 ) Pub Date : 2018-09-14 00:00:00 , DOI: 10.1021/acs.jchemed.8b00225 Howard Mayes 1 , Chung F. Wong 1, 2
Affiliation
|
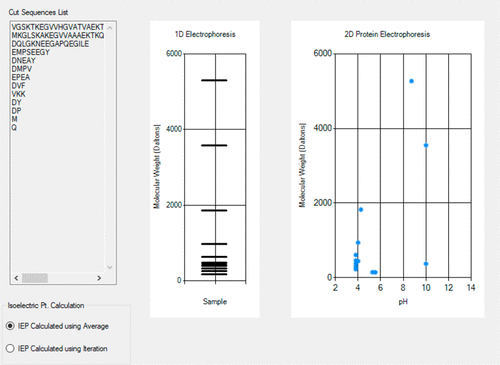
A new program, ECEP2D, for simulating the one-dimensional (1D) and two-dimensional (2D) patterns of the gel electrophoresis of a protein after it has been digested by one or more enzymes is introduced. With ECEP2D, students can gain deeper insights into gel electrophoresis by performing hands-on simulations. For example, students can visualize how 2D gel electrophoresis can improve resolution over 1D by comparing them side-by-side. Students can watch how different enzymes cut a protein into different fragments, giving rise to different gel patterns; some patterns are less congested than the others. Students can recognize how enzyme digestion can enhance gel electrophoresis to distinguish proteins. For students not having the chance to take biochemistry laboratories, ECEP2D provides a useful simulated learning environment. Instructors can also complement wet-lab experiments with simulations by ECEP2D to introduce more concepts with fewer laboratory hours. ECEP2D is available for free at http://www.umsl.edu/~wongch/software.html.
中文翻译:

酶消化蛋白的凝胶电泳模拟程序
引入了新程序ECEP2D,用于模拟蛋白质被一种或多种酶消化后的凝胶电泳的一维(1D)和二维(2D)模式。借助ECEP2D,学生可以通过执行动手模拟来更深入地了解凝胶电泳。例如,学生可以通过并排比较2D凝胶电泳与1D相比如何改善分辨率。学生可以观察到不同的酶如何将蛋白质切割成不同的片段,从而产生不同的凝胶模式。一些模式比其他模式少拥挤。学生可以认识到酶消化如何增强凝胶电泳来区分蛋白质。对于没有机会参加生物化学实验室学习的学生,ECEP2D提供了有用的模拟学习环境。讲师还可以通过ECEP2D的模拟来补充湿实验室实验,从而以更少的实验室时间介绍更多的概念。ECEP2D可从http://www.umsl.edu/~wongch/software.html免费获得。
更新日期:2018-09-14
中文翻译:

酶消化蛋白的凝胶电泳模拟程序
引入了新程序ECEP2D,用于模拟蛋白质被一种或多种酶消化后的凝胶电泳的一维(1D)和二维(2D)模式。借助ECEP2D,学生可以通过执行动手模拟来更深入地了解凝胶电泳。例如,学生可以通过并排比较2D凝胶电泳与1D相比如何改善分辨率。学生可以观察到不同的酶如何将蛋白质切割成不同的片段,从而产生不同的凝胶模式。一些模式比其他模式少拥挤。学生可以认识到酶消化如何增强凝胶电泳来区分蛋白质。对于没有机会参加生物化学实验室学习的学生,ECEP2D提供了有用的模拟学习环境。讲师还可以通过ECEP2D的模拟来补充湿实验室实验,从而以更少的实验室时间介绍更多的概念。ECEP2D可从http://www.umsl.edu/~wongch/software.html免费获得。











































 京公网安备 11010802027423号
京公网安备 11010802027423号