当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Design of Large-Scale Reporter Construct Arrays for Dynamic, Live Cell Systems Biology
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-09-06 00:00:00 , DOI: 10.1021/acssynbio.8b00236 Joseph T Decker 1 , Matthew S Hall 1 , Beatriz Peñalver-Bernabé 2 , Rachel B Blaisdell 1 , Lauren N Liebman 1 , Jacqueline S Jeruss 1, 3 , Lonnie D Shea 1
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2018-09-06 00:00:00 , DOI: 10.1021/acssynbio.8b00236 Joseph T Decker 1 , Matthew S Hall 1 , Beatriz Peñalver-Bernabé 2 , Rachel B Blaisdell 1 , Lauren N Liebman 1 , Jacqueline S Jeruss 1, 3 , Lonnie D Shea 1
Affiliation
|
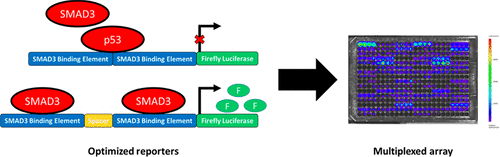
Dynamic systems biology aims to identify the molecular mechanisms governing cell fate decisions through the analysis of living cells. Large scale molecular information from living cells can be obtained from reporter constructs that provide activities for either individual transcription factors or multiple factors binding to the full promoter following CRISPR/Cas9 directed insertion of luciferase. In this report, we investigated the design criteria to obtain reporters that are specific and responsive to transcription factor (TF) binding and the integration of TF binding activity with genetic reporter activity. The design of TF reporters was investigated for the impact of consensus binding site spacing sequence and off-target binding on the reporter sensitivity using a library of 25 SMAD3 activity reporters with spacers of random composition and length. A spacer was necessary to quantify activity changes after TGFβ stimulation. TF binding site prediction algorithms (BEEML, FIMO and DeepBind) were used to predict off-target binding, and nonresponsiveness to a SMAD3 reporter was correlated with a predicted competitive binding of constitutively active p53. The network of activity of the SMAD3 reporter was inferred from measurements of TF reporter library, and connected with large-scale genetic reporter activity measurements. The integration of TF and genetic reporters identified the major hubs directing responses to TGFβ, and this method provided a systems-level algorithm to investigate cell signaling.
中文翻译:

用于动态活细胞系统生物学的大规模报告构建体阵列的设计
动态系统生物学旨在通过分析活细胞来确定控制细胞命运决定的分子机制。来自活细胞的大规模分子信息可以从报告构建体中获得,该报告构建体为单个转录因子或在 CRISPR/Cas9 定向插入荧光素酶后与完整启动子结合的多个因子提供活性。在本报告中,我们研究了设计标准,以获得对转录因子 (TF) 结合具有特异性和响应性的报告基因,以及 TF 结合活性与遗传报告基因活性的整合。使用包含随机组成和长度间隔的 25 个 SMAD3 活性报告基因库,研究了 TF 报告基因的设计,以了解共有结合位点间隔序列和脱靶结合对报告基因敏感性的影响。需要使用间隔物来量化 TGFβ 刺激后的活性变化。 TF 结合位点预测算法(BEEML、FIMO 和 DeepBind)用于预测脱靶结合,并且对 SMAD3 报告基因的无反应性与预测的组成型活性 p53 的竞争性结合相关。 SMAD3 报告基因的活性网络是根据 TF 报告基因库的测量推断出来的,并与大规模遗传报告基因活性测量相联系。 TF 和遗传报告基因的整合确定了指导 TGFβ 反应的主要中枢,并且该方法提供了系统级算法来研究细胞信号传导。
更新日期:2018-09-06
中文翻译:

用于动态活细胞系统生物学的大规模报告构建体阵列的设计
动态系统生物学旨在通过分析活细胞来确定控制细胞命运决定的分子机制。来自活细胞的大规模分子信息可以从报告构建体中获得,该报告构建体为单个转录因子或在 CRISPR/Cas9 定向插入荧光素酶后与完整启动子结合的多个因子提供活性。在本报告中,我们研究了设计标准,以获得对转录因子 (TF) 结合具有特异性和响应性的报告基因,以及 TF 结合活性与遗传报告基因活性的整合。使用包含随机组成和长度间隔的 25 个 SMAD3 活性报告基因库,研究了 TF 报告基因的设计,以了解共有结合位点间隔序列和脱靶结合对报告基因敏感性的影响。需要使用间隔物来量化 TGFβ 刺激后的活性变化。 TF 结合位点预测算法(BEEML、FIMO 和 DeepBind)用于预测脱靶结合,并且对 SMAD3 报告基因的无反应性与预测的组成型活性 p53 的竞争性结合相关。 SMAD3 报告基因的活性网络是根据 TF 报告基因库的测量推断出来的,并与大规模遗传报告基因活性测量相联系。 TF 和遗传报告基因的整合确定了指导 TGFβ 反应的主要中枢,并且该方法提供了系统级算法来研究细胞信号传导。











































 京公网安备 11010802027423号
京公网安备 11010802027423号