当前位置:
X-MOL 学术
›
Mol. Pharmaceutics
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Entangled Conditional Adversarial Autoencoder for de Novo Drug Discovery
Molecular Pharmaceutics ( IF 4.5 ) Pub Date : 2018-09-04 00:00:00 , DOI: 10.1021/acs.molpharmaceut.8b00839 Daniil Polykovskiy,Alexander Zhebrak,Dmitry Vetrov,Yan Ivanenkov,Vladimir Aladinskiy,Polina Mamoshina,Marine Bozdaganyan,Alexander Aliper,Alex Zhavoronkov,Artur Kadurin
Molecular Pharmaceutics ( IF 4.5 ) Pub Date : 2018-09-04 00:00:00 , DOI: 10.1021/acs.molpharmaceut.8b00839 Daniil Polykovskiy,Alexander Zhebrak,Dmitry Vetrov,Yan Ivanenkov,Vladimir Aladinskiy,Polina Mamoshina,Marine Bozdaganyan,Alexander Aliper,Alex Zhavoronkov,Artur Kadurin
|
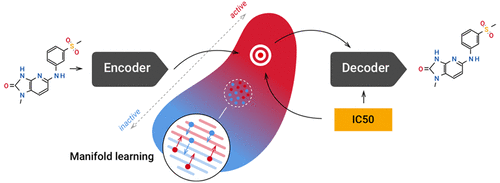
Modern computational approaches and machine learning techniques accelerate the invention of new drugs. Generative models can discover novel molecular structures within hours, while conventional drug discovery pipelines require months of work. In this article, we propose a new generative architecture, entangled conditional adversarial autoencoder, that generates molecular structures based on various properties, such as activity against a specific protein, solubility, or ease of synthesis. We apply the proposed model to generate a novel inhibitor of Janus kinase 3, implicated in rheumatoid arthritis, psoriasis, and vitiligo. The discovered molecule was tested in vitro and showed good activity and selectivity.
中文翻译:

纠缠的有条件对抗自动编码器,用于从头发现药物
现代计算方法和机器学习技术加速了新药的发明。生成模型可以在数小时内发现新颖的分子结构,而传统的药物发现流程则需要数月的工作。在本文中,我们提出了一种新的生成体系结构,即纠缠的条件对抗自动编码器,它可以基于各种特性(例如针对特定蛋白质的活性,溶解性或易于合成)生成分子结构。我们应用提出的模型来产生新型的Janus激酶抑制剂3,涉及类风湿性关节炎,牛皮癣和白癜风。对发现的分子进行了体外测试,显示出良好的活性和选择性。
更新日期:2018-09-04
中文翻译:

纠缠的有条件对抗自动编码器,用于从头发现药物
现代计算方法和机器学习技术加速了新药的发明。生成模型可以在数小时内发现新颖的分子结构,而传统的药物发现流程则需要数月的工作。在本文中,我们提出了一种新的生成体系结构,即纠缠的条件对抗自动编码器,它可以基于各种特性(例如针对特定蛋白质的活性,溶解性或易于合成)生成分子结构。我们应用提出的模型来产生新型的Janus激酶抑制剂3,涉及类风湿性关节炎,牛皮癣和白癜风。对发现的分子进行了体外测试,显示出良好的活性和选择性。











































 京公网安备 11010802027423号
京公网安备 11010802027423号