当前位置:
X-MOL 学术
›
Cell Chem. Bio.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Translational Reprogramming Provides a Blueprint for Cellular Adaptation
Cell Chemical Biology ( IF 6.6 ) Pub Date : 2018-08-30 , DOI: 10.1016/j.chembiol.2018.08.003 Max Berman Ferretti , Jennifer Louise Barre , Katrin Karbstein
Cell Chemical Biology ( IF 6.6 ) Pub Date : 2018-08-30 , DOI: 10.1016/j.chembiol.2018.08.003 Max Berman Ferretti , Jennifer Louise Barre , Katrin Karbstein
|
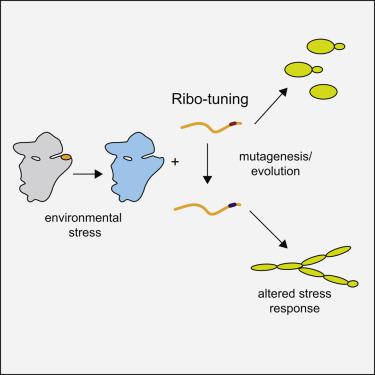
Consistent with its location on the ribosome, reporter assays demonstrate a role for Rps26 in recognition of the Kozak sequence. Consequently, Rps26-deficient ribosomes display preference for mRNAs encoding components of the high salt and high pH stress response pathways and accumulate in yeast exposed to high salt or pH. Here we use this information to reprogram the cellular response to high salt by introducing point mutations in the Kozak sequence of key regulators for the cell wall MAP-kinase, filamentation, or DNA repair pathways. This stimulates their translation upon genetic, or salt-induced Rps26 depletion from ribosomes. Stress resistance assays show activation of the targeted pathways in an Rps26- and salt-dependent manner. Genomic alterations in diverse yeast populations indicate that analogous tuning occurs during adaptation to ecological niches. Thus, evolution shapes translational control across the genome by taking advantage of the accumulation of diverse ribosome populations.
中文翻译:

翻译重编程为细胞适应提供了蓝图
与它在核糖体上的位置一致,报告基因检测证明Rps26在识别Kozak序列中的作用。因此,缺乏Rps26的核糖体显示出对编码高盐和高pH应激反应途径成分的mRNA的偏好,并在暴露于高盐或pH的酵母中积累。在这里,我们通过在细胞壁MAP激酶,细丝化或DNA修复途径的关键调控因子的Kozak序列中引入点突变,利用这些信息来重新编程对高盐的细胞应答。当遗传或盐诱导的核糖体耗尽Rps26时,这会刺激它们的翻译。压力抗性测定显示以Rps26和盐依赖性方式激活目标途径。不同酵母种群的基因组变化表明,在适应生态位的过程中发生了类似的调节。因此,进化通过利用各种核糖体群体的积累来塑造整个基因组的翻译控制。
更新日期:2018-11-15
中文翻译:

翻译重编程为细胞适应提供了蓝图
与它在核糖体上的位置一致,报告基因检测证明Rps26在识别Kozak序列中的作用。因此,缺乏Rps26的核糖体显示出对编码高盐和高pH应激反应途径成分的mRNA的偏好,并在暴露于高盐或pH的酵母中积累。在这里,我们通过在细胞壁MAP激酶,细丝化或DNA修复途径的关键调控因子的Kozak序列中引入点突变,利用这些信息来重新编程对高盐的细胞应答。当遗传或盐诱导的核糖体耗尽Rps26时,这会刺激它们的翻译。压力抗性测定显示以Rps26和盐依赖性方式激活目标途径。不同酵母种群的基因组变化表明,在适应生态位的过程中发生了类似的调节。因此,进化通过利用各种核糖体群体的积累来塑造整个基因组的翻译控制。











































 京公网安备 11010802027423号
京公网安备 11010802027423号