Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling
Nature ( IF 50.5 ) Pub Date : 2018-08-29 , DOI: 10.1038/s41586-018-0462-y Ayala Shiber 1, 2 , Kristina Döring 1, 2 , Ulrike Friedrich 1, 2 , Kevin Klann 1, 2 , Dorina Merker 1, 2 , Mostafa Zedan 1, 2 , Frank Tippmann 1, 2 , Günter Kramer 1, 2 , Bernd Bukau 1, 2
Nature ( IF 50.5 ) Pub Date : 2018-08-29 , DOI: 10.1038/s41586-018-0462-y Ayala Shiber 1, 2 , Kristina Döring 1, 2 , Ulrike Friedrich 1, 2 , Kevin Klann 1, 2 , Dorina Merker 1, 2 , Mostafa Zedan 1, 2 , Frank Tippmann 1, 2 , Günter Kramer 1, 2 , Bernd Bukau 1, 2
Affiliation
|
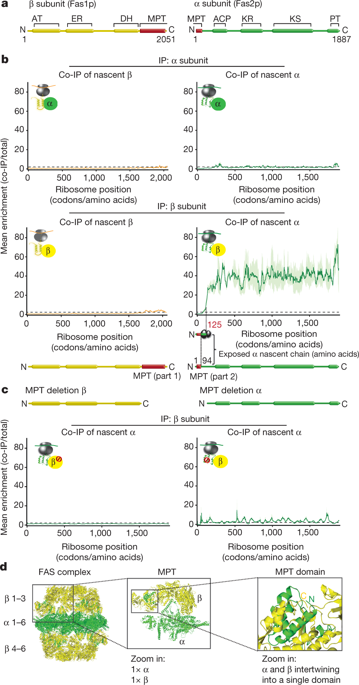
The folding of newly synthesized proteins to the native state is a major challenge within the crowded cellular environment, as non-productive interactions can lead to misfolding, aggregation and degradation1. Cells cope with this challenge by coupling synthesis with polypeptide folding and by using molecular chaperones to safeguard folding cotranslationally2. However, although most of the cellular proteome forms oligomeric assemblies3, little is known about the final step of folding: the assembly of polypeptides into complexes. In prokaryotes, a proof-of-concept study showed that the assembly of heterodimeric luciferase is an organized cotranslational process that is facilitated by spatially confined translation of the subunits encoded on a polycistronic mRNA4. In eukaryotes, however, fundamental differences—such as the rarity of polycistronic mRNAs and different chaperone constellations—raise the question of whether assembly is also coordinated with translation. Here we provide a systematic and mechanistic analysis of the assembly of protein complexes in eukaryotes using ribosome profiling. We determined the in vivo interactions of the nascent subunits from twelve hetero-oligomeric protein complexes of Saccharomyces cerevisiae at near-residue resolution. We find nine complexes assemble cotranslationally; the three complexes that do not show cotranslational interactions are regulated by dedicated assembly chaperones5–7. Cotranslational assembly often occurs uni-directionally, with one fully synthesized subunit engaging its nascent partner subunit, thereby counteracting its propensity for aggregation. The onset of cotranslational subunit association coincides directly with the full exposure of the nascent interaction domain at the ribosomal tunnel exit. The action of the ribosome-associated Hsp70 chaperone Ssb8 is coordinated with assembly. Ssb transiently engages partially synthesized interaction domains and then dissociates before the onset of partner subunit association, presumably to prevent premature assembly interactions. Our study shows that cotranslational subunit association is a prevalent mechanism for the assembly of hetero-oligomers in yeast and indicates that translation, folding and the assembly of protein complexes are integrated processes in eukaryotes.Cotranslational assembly is a prevalent mechanism for the formation of oligomeric complexes in Saccharomyces cerevisiae, with one subunit serving as scaffold for the translation of partner subunits.
中文翻译:

核糖体分析揭示真核生物中蛋白质复合物的共翻译组装
在拥挤的细胞环境中,将新合成的蛋白质折叠成天然状态是一项重大挑战,因为非生产性相互作用会导致错误折叠、聚集和降解1。细胞通过将合成与多肽折叠结合并使用分子伴侣来保护折叠共翻译来应对这一挑战。然而,尽管大多数细胞蛋白质组形成寡聚组装体3,但人们对折叠的最后一步知之甚少:将多肽组装成复合物。在原核生物中,一项概念验证研究表明,异二聚体荧光素酶的组装是一个有组织的共翻译过程,可通过多顺反子 mRNA4 上编码的亚基的空间受限翻译来促进。然而,在真核生物中,基本的差异——例如多顺反子 mRNA 的稀有性和不同的伴侣星座——提出了组装是否也与翻译相协调的问题。在这里,我们使用核糖体分析对真核生物中蛋白质复合物的组装进行了系统和机械分析。我们确定了来自酿酒酵母的 12 个异源寡聚蛋白复合物的新生亚基在近残留分辨率下的体内相互作用。我们发现九个复合体以共翻译方式组装;不显示共翻译相互作用的三个复合物由专门的组装伴侣调节 5-7。共翻译组装通常单向发生,一个完全合成的亚基与其新生的伙伴亚基接合,从而抵消了其聚集的倾向。共翻译亚基结合的开始与核糖体隧道出口处新生相互作用域的完全暴露直接一致。核糖体相关的 Hsp70 伴侣 Ssb8 的作用与组装相协调。Ssb 短暂地参与部分合成的相互作用域,然后在伙伴亚基结合开始之前解离,大概是为了防止过早的组装相互作用。我们的研究表明,共翻译亚基结合是酵母中异源寡聚体组装的普遍机制,表明蛋白质复合物的翻译、折叠和组装是真核生物中的整合过程。共翻译组装是寡聚复合物形成的普遍机制在酿酒酵母中,
更新日期:2018-08-29
中文翻译:

核糖体分析揭示真核生物中蛋白质复合物的共翻译组装
在拥挤的细胞环境中,将新合成的蛋白质折叠成天然状态是一项重大挑战,因为非生产性相互作用会导致错误折叠、聚集和降解1。细胞通过将合成与多肽折叠结合并使用分子伴侣来保护折叠共翻译来应对这一挑战。然而,尽管大多数细胞蛋白质组形成寡聚组装体3,但人们对折叠的最后一步知之甚少:将多肽组装成复合物。在原核生物中,一项概念验证研究表明,异二聚体荧光素酶的组装是一个有组织的共翻译过程,可通过多顺反子 mRNA4 上编码的亚基的空间受限翻译来促进。然而,在真核生物中,基本的差异——例如多顺反子 mRNA 的稀有性和不同的伴侣星座——提出了组装是否也与翻译相协调的问题。在这里,我们使用核糖体分析对真核生物中蛋白质复合物的组装进行了系统和机械分析。我们确定了来自酿酒酵母的 12 个异源寡聚蛋白复合物的新生亚基在近残留分辨率下的体内相互作用。我们发现九个复合体以共翻译方式组装;不显示共翻译相互作用的三个复合物由专门的组装伴侣调节 5-7。共翻译组装通常单向发生,一个完全合成的亚基与其新生的伙伴亚基接合,从而抵消了其聚集的倾向。共翻译亚基结合的开始与核糖体隧道出口处新生相互作用域的完全暴露直接一致。核糖体相关的 Hsp70 伴侣 Ssb8 的作用与组装相协调。Ssb 短暂地参与部分合成的相互作用域,然后在伙伴亚基结合开始之前解离,大概是为了防止过早的组装相互作用。我们的研究表明,共翻译亚基结合是酵母中异源寡聚体组装的普遍机制,表明蛋白质复合物的翻译、折叠和组装是真核生物中的整合过程。共翻译组装是寡聚复合物形成的普遍机制在酿酒酵母中,











































 京公网安备 11010802027423号
京公网安备 11010802027423号