当前位置:
X-MOL 学术
›
Cell Chem. Bio.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Combining Mutational Signatures, Clonal Fitness, and Drug Affinity to Define Drug-Specific Resistance Mutations in Cancer
Cell Chemical Biology ( IF 6.6 ) Pub Date : 2018-08-23 , DOI: 10.1016/j.chembiol.2018.07.013 Teresa Kaserer 1 , Julian Blagg 1
Cell Chemical Biology ( IF 6.6 ) Pub Date : 2018-08-23 , DOI: 10.1016/j.chembiol.2018.07.013 Teresa Kaserer 1 , Julian Blagg 1
Affiliation
|
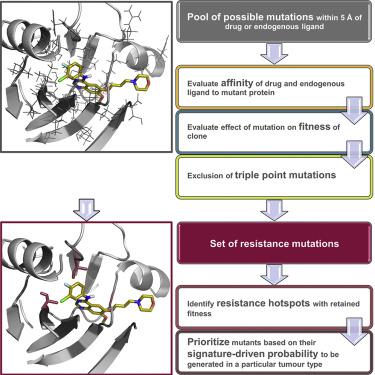
The emergence of mutations that confer resistance to molecularly targeted therapeutics is dependent upon the effect of each mutation on drug affinity for the target protein, the clonal fitness of cells harboring the mutation, and the probability that each variant can be generated by DNA codon base mutation. We present a computational workflow that combines these three factors to identify mutations likely to arise upon drug treatment in a particular tumor type. The Osprey-based workflow is validated using a comprehensive dataset of ERK2 mutations and is applied to small-molecule drugs and/or therapeutic antibodies targeting KIT, EGFR, Abl, and ALK. We identify major clinically observed drug-resistant mutations for drug-target pairs and highlight the potential to prospectively identify probable drug resistance mutations.
中文翻译:

结合突变特征、克隆适应性和药物亲和力来定义癌症中的药物特异性耐药突变
赋予分子靶向治疗耐药性的突变的出现取决于每个突变对靶蛋白药物亲和力的影响、携带突变的细胞的克隆适应性以及每个变体可由 DNA 密码子碱基突变产生的概率。我们提出了一个计算工作流程,结合这三个因素来识别特定肿瘤类型的药物治疗时可能出现的突变。基于 Osprey 的工作流程使用 ERK2 突变的综合数据集进行验证,并应用于针对 KIT、EGFR、Abl 和 ALK 的小分子药物和/或治疗抗体。我们确定了临床观察到的药物靶点对的主要耐药突变,并强调了前瞻性识别可能的耐药突变的潜力。
更新日期:2018-11-15
中文翻译:

结合突变特征、克隆适应性和药物亲和力来定义癌症中的药物特异性耐药突变
赋予分子靶向治疗耐药性的突变的出现取决于每个突变对靶蛋白药物亲和力的影响、携带突变的细胞的克隆适应性以及每个变体可由 DNA 密码子碱基突变产生的概率。我们提出了一个计算工作流程,结合这三个因素来识别特定肿瘤类型的药物治疗时可能出现的突变。基于 Osprey 的工作流程使用 ERK2 突变的综合数据集进行验证,并应用于针对 KIT、EGFR、Abl 和 ALK 的小分子药物和/或治疗抗体。我们确定了临床观察到的药物靶点对的主要耐药突变,并强调了前瞻性识别可能的耐药突变的潜力。











































 京公网安备 11010802027423号
京公网安备 11010802027423号