当前位置:
X-MOL 学术
›
Microchem. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Classification of Paris species according to botanical and geographical origins based on spectroscopic, chromatographic, conventional chemometric analysis and data fusion strategy
Microchemical Journal ( IF 4.9 ) Pub Date : 2018-12-01 , DOI: 10.1016/j.microc.2018.08.035 Xue-Mei Wu , Zhi-Tian Zuo , Qing-Zhi Zhang , Yuan-Zhong Wang
Microchemical Journal ( IF 4.9 ) Pub Date : 2018-12-01 , DOI: 10.1016/j.microc.2018.08.035 Xue-Mei Wu , Zhi-Tian Zuo , Qing-Zhi Zhang , Yuan-Zhong Wang
|
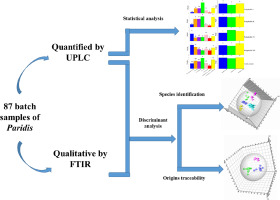
Abstract Given that the consumption of pure varieties for Paridis Rhizome was increasing and causing a “ripple effect”, i.e. numerous other unofficial Paris species were harvested and traded as the succedaneum of Paridis. The purpose of this study was to establish an effective strategy to distinguish Paris species and trace geographical origins of pure varieties of Paridis samples. A total of 87 batch samples of wild Paris were analyzed by UPLC and FTIR. Among them, 79 batch samples of five species and 40 batch samples of Paris polyphylla Smith var. yunnanensis (Franch.) Hand.-Mazz were used to establish pattern recognition models of PLS-DA, SVM and random forest simultaneously, basing on polyphyllin content, FTIR spectra, UPLC chromatograms, low- and mid-level data fusion strategy. Quantitative analysis results revealed that more obvious fluctuation of polyphyllin content was shown in different species sample than that of pure varieties of Paridis collected from various origins. Pattern recognition models showed that compared with polyphyllin content, FTIR spectra, UPLC chromatograms and low-level data fusion method, mid-level data fusion strategy showed a better accuracy (>92%) in PLS-DA model to classify Paridis samples according to botanical and geographical origins. Similar results were obtained in SVM and random forest models. Thus, the classification of these three pattern recognition models were verified each other and demonstrated that mid-level data fusion strategy combined with chemometrics could recognize different Paris species and trace the pure varieties of Paridis geographical origins, correctly.
中文翻译:

基于光谱、色谱、常规化学计量分析和数据融合策略,根据植物和地理起源对巴黎物种进行分类
摘要 鉴于Paridis Rhizome 纯品种的消费量不断增加并引起“涟漪效应”,即大量其他非官方Paridis 物种被收获并作为Paridis 的继承物进行交易。本研究的目的是建立一种有效的策略来区分巴黎物种并追踪 Paridis 样本纯品种的地理起源。通过 UPLC 和 FTIR 分析了总共 87 批野生巴黎的样品。其中,5种79批次样品,巴黎多叶史密斯变种40批次样品。基于多叶素含量、FTIR光谱、UPLC色谱图、中低级数据融合策略,利用云南省汉德.-Mazz同时建立PLS-DA、SVM和随机森林的模式识别模型。定量分析结果表明,与不同产地采集的纯种凤仙花相比,不同物种样品中多叶素含量的波动更为明显。模式识别模型表明,与多叶素含量、FTIR 光谱、UPLC 色谱图和低级数据融合方法相比,中级数据融合策略在 PLS-DA 模型中显示出更好的准确率(>92%),根据植物对 Paridis 样品进行分类和地理起源。在 SVM 和随机森林模型中获得了类似的结果。因此,这三种模式识别模型的分类相互验证,表明结合化学计量学的中级数据融合策略可以正确识别不同的巴黎物种,并正确追溯Paridis地理起源的纯品种。
更新日期:2018-12-01
中文翻译:

基于光谱、色谱、常规化学计量分析和数据融合策略,根据植物和地理起源对巴黎物种进行分类
摘要 鉴于Paridis Rhizome 纯品种的消费量不断增加并引起“涟漪效应”,即大量其他非官方Paridis 物种被收获并作为Paridis 的继承物进行交易。本研究的目的是建立一种有效的策略来区分巴黎物种并追踪 Paridis 样本纯品种的地理起源。通过 UPLC 和 FTIR 分析了总共 87 批野生巴黎的样品。其中,5种79批次样品,巴黎多叶史密斯变种40批次样品。基于多叶素含量、FTIR光谱、UPLC色谱图、中低级数据融合策略,利用云南省汉德.-Mazz同时建立PLS-DA、SVM和随机森林的模式识别模型。定量分析结果表明,与不同产地采集的纯种凤仙花相比,不同物种样品中多叶素含量的波动更为明显。模式识别模型表明,与多叶素含量、FTIR 光谱、UPLC 色谱图和低级数据融合方法相比,中级数据融合策略在 PLS-DA 模型中显示出更好的准确率(>92%),根据植物对 Paridis 样品进行分类和地理起源。在 SVM 和随机森林模型中获得了类似的结果。因此,这三种模式识别模型的分类相互验证,表明结合化学计量学的中级数据融合策略可以正确识别不同的巴黎物种,并正确追溯Paridis地理起源的纯品种。











































 京公网安备 11010802027423号
京公网安备 11010802027423号