当前位置:
X-MOL 学术
›
ACS Infect. Dis.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Direct-qPCR Assay for Coupled Identification and Antimicrobial Susceptibility Testing of Neisseria gonorrhoeae
ACS Infectious Diseases ( IF 4.0 ) Pub Date : 2018-07-12 00:00:00 , DOI: 10.1021/acsinfecdis.8b00104 Liben Chen , Dong Jin Shin , Shuyu Zheng , Johan H. Melendez 1 , Charlotte A. Gaydos 1 , Tza-Huei Wang
ACS Infectious Diseases ( IF 4.0 ) Pub Date : 2018-07-12 00:00:00 , DOI: 10.1021/acsinfecdis.8b00104 Liben Chen , Dong Jin Shin , Shuyu Zheng , Johan H. Melendez 1 , Charlotte A. Gaydos 1 , Tza-Huei Wang
Affiliation
|
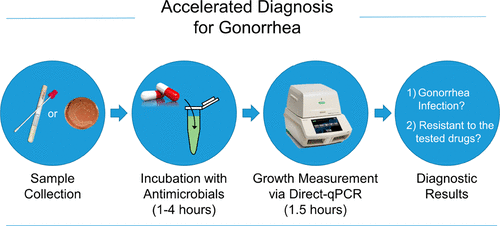
Multidrug-resistant gonorrhea has become an urgent issue for global public health. As the causative agent of gonorrhea, Neisseria gonorrhoeae, has been progressively developing resistance to nearly all prescribed antimicrobial drugs, monitoring its antimicrobial resistance on a broader scale has become a crucial agenda for effective antibiotic stewardship. Unfortunately, gold standard antimicrobial susceptibility testing (AST) relies on time and labor-intensive phenotypic assays, which lag behind the current diagnostic workflow for N. gonorrhoeae identification based on nucleic acid amplification tests (NAAT). Newer assay technologies based on NAAT can rapidly identify N. gonorrhoeae from clinical specimen but fundamentally lack the capacity to provide phenotypic AST information. Herein, we propose a direct-quantitative PCR (direct-qPCR) assay that enables pathogen-specific identification and phenotypic AST via quantitative measurement of N. gonorrhoeae growth directly from a liquid medium without any sample preprocessing. The assay has an analytical sensitivity of 102 CFU/mL and is highly specific to N. gonorrhoeae in the presence of urogenital flora and clinical swab eluent. We tested seven N. gonorrhoeae strains against three antibiotic agents, penicillin, tetracycline, and ciprofloxacin, and achieved 95.2% category agreement and 85.7% essential agreement with the FDA-approved E-test. The assay presented in this work has the unique ability to identify N. gonorrhoeae and provide phenotypic AST directly from the liquid medium with cell densities as low as 102 CFU/mL, demonstrating an accelerated, sensitive, and scalable workflow for performing both identification and AST of N. gonorrhoeae.
中文翻译:

Direct-qPCR分析法用于淋病奈瑟菌的鉴定和抗菌药敏试验
耐多药淋病已成为全球公共卫生的紧迫问题。作为淋病的病原体,淋病奈瑟氏球菌已逐渐发展出对几乎所有处方抗菌药物的耐药性,在更广泛的规模上监测其耐药性已成为有效抗生素管理的重要议程。不幸的是,金标准的抗生素敏感性测试(AST)依赖于时间和劳动强度大的表型分析,这落后于当前基于核酸扩增测试(NAAT)鉴定淋病奈瑟氏球菌的诊断工作流程。基于NAAT的更新检测技术可以快速鉴定淋病奈瑟氏球菌从临床标本中提取,但从根本上来说无法提供表型AST信息。本文中,我们提出了一种直接定量PCR(direct-qPCR)分析方法,该方法可通过直接从液体培养基定量测定淋病奈瑟氏球菌而无需进行任何样品预处理,即可进行病原体特异性鉴定和表型AST 。该方法的分析灵敏度为10 2 CFU / mL,在存在泌尿生殖道菌群和临床拭子洗脱液的情况下对淋病奈瑟菌具有高度特异性。我们测试了七个淋病奈瑟氏球菌菌株针对三种抗生素药物:青霉素,四环素和环丙沙星,通过FDA批准的E-test达到95.2%的类别一致性和85.7%的基本一致性。这项工作中介绍的测定法具有独特的能力来鉴定淋病奈瑟氏球菌,并直接从细胞密度低至10 2 CFU / mL的液体培养基中提供表型AST ,证明了进行鉴定和鉴定的加速,灵敏和可扩展的工作流程淋病奈瑟氏球菌的AST 。
更新日期:2018-07-12
中文翻译:

Direct-qPCR分析法用于淋病奈瑟菌的鉴定和抗菌药敏试验
耐多药淋病已成为全球公共卫生的紧迫问题。作为淋病的病原体,淋病奈瑟氏球菌已逐渐发展出对几乎所有处方抗菌药物的耐药性,在更广泛的规模上监测其耐药性已成为有效抗生素管理的重要议程。不幸的是,金标准的抗生素敏感性测试(AST)依赖于时间和劳动强度大的表型分析,这落后于当前基于核酸扩增测试(NAAT)鉴定淋病奈瑟氏球菌的诊断工作流程。基于NAAT的更新检测技术可以快速鉴定淋病奈瑟氏球菌从临床标本中提取,但从根本上来说无法提供表型AST信息。本文中,我们提出了一种直接定量PCR(direct-qPCR)分析方法,该方法可通过直接从液体培养基定量测定淋病奈瑟氏球菌而无需进行任何样品预处理,即可进行病原体特异性鉴定和表型AST 。该方法的分析灵敏度为10 2 CFU / mL,在存在泌尿生殖道菌群和临床拭子洗脱液的情况下对淋病奈瑟菌具有高度特异性。我们测试了七个淋病奈瑟氏球菌菌株针对三种抗生素药物:青霉素,四环素和环丙沙星,通过FDA批准的E-test达到95.2%的类别一致性和85.7%的基本一致性。这项工作中介绍的测定法具有独特的能力来鉴定淋病奈瑟氏球菌,并直接从细胞密度低至10 2 CFU / mL的液体培养基中提供表型AST ,证明了进行鉴定和鉴定的加速,灵敏和可扩展的工作流程淋病奈瑟氏球菌的AST 。











































 京公网安备 11010802027423号
京公网安备 11010802027423号