当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
In Vitro Transcription Networks Based on Hairpin Promoter Switches
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-07-18 00:00:00 , DOI: 10.1021/acssynbio.8b00172 Shaunak Kar , Andrew D. Ellington
ACS Synthetic Biology ( IF 4.7 ) Pub Date : 2018-07-18 00:00:00 , DOI: 10.1021/acssynbio.8b00172 Shaunak Kar , Andrew D. Ellington
|
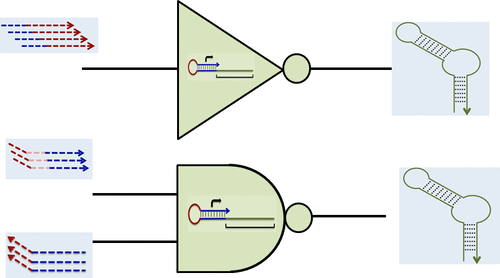
In vitro transcription networks are analogs of naturally occurring gene regulatory networks that consist of synthetic DNA templates that are cross-regulated by their own transcripts. This ability to design and execute in vitro transcription networks has allowed bottom-up construction of complex network topologies with predictable dynamic behavior. Here we describe the simplified design of an in vitro transcription network based on single-stranded synthetic DNA hairpin switches that function similar to molecular beacons, via toehold mediated strand displacement. Systematic construction of increasingly larger circuits was achieved by programming interactions between multiple switches through rational sequence design, and the dynamic behavior of networks was accurately predicted using a simple mathematical model. Ultimately, we engineered a cascade of switches that acted as a Boolean complete NAND gate capable of sensing both DNA and RNA inputs. The tools and framework that have been developed makes the execution of in vitro transcription circuits much simpler, which will enable them to more readily serve as testbeds for nucleic acid computations both in vitro and in vivo.
中文翻译:

基于发夹启动子开关的体外转录网络
体外转录网络是天然存在的基因调控网络的类似物,该基因调控网络由合成的DNA模板组成,这些模板被其自身的转录物交叉调控。设计和执行体外转录网络的能力允许自底向上构建具有可预测动态行为的复杂网络拓扑。在这里,我们描述了基于单链合成DNA发夹开关的体外转录网络的简化设计,该功能类似于分子信标,通过脚趾介导的链移位。通过合理的序列设计对多个开关之间的交互进行编程,可以实现越来越大的电路的系统构建,并且可以使用简单的数学模型准确预测网络的动态行为。最终,我们设计了一系列级联的开关,它们充当了能够感应DNA和RNA输入的布尔型完整NAND门。已经开发的工具和框架使体外转录电路的执行变得更加简单,这将使它们能够更容易地用作体外和体内核酸计算的测试平台。
更新日期:2018-07-18
中文翻译:

基于发夹启动子开关的体外转录网络
体外转录网络是天然存在的基因调控网络的类似物,该基因调控网络由合成的DNA模板组成,这些模板被其自身的转录物交叉调控。设计和执行体外转录网络的能力允许自底向上构建具有可预测动态行为的复杂网络拓扑。在这里,我们描述了基于单链合成DNA发夹开关的体外转录网络的简化设计,该功能类似于分子信标,通过脚趾介导的链移位。通过合理的序列设计对多个开关之间的交互进行编程,可以实现越来越大的电路的系统构建,并且可以使用简单的数学模型准确预测网络的动态行为。最终,我们设计了一系列级联的开关,它们充当了能够感应DNA和RNA输入的布尔型完整NAND门。已经开发的工具和框架使体外转录电路的执行变得更加简单,这将使它们能够更容易地用作体外和体内核酸计算的测试平台。



























 京公网安备 11010802027423号
京公网安备 11010802027423号