当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Highly Ordered and Field-free 3D DNA Nanostructure: The Next Generation of DNA Nanomachine for Rapid Single-step Sensing
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2018-07-16 , DOI: 10.1021/jacs.8b04648 Pu Zhang 1 , Jie Jiang 1 , Ruo Yuan 1 , Ying Zhuo 1 , Yaqin Chai 1
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2018-07-16 , DOI: 10.1021/jacs.8b04648 Pu Zhang 1 , Jie Jiang 1 , Ruo Yuan 1 , Ying Zhuo 1 , Yaqin Chai 1
Affiliation
|
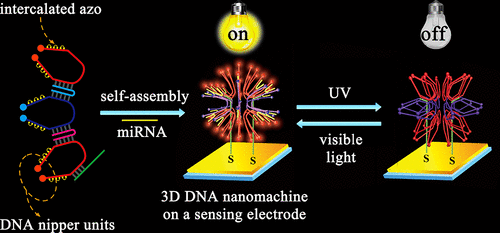
Herein, by directly using Watson-Crick base pairing, a highly ordered and field-free three-dimensional (3D) DNA nanostructure is self-assembled by azobenzene (azo)-functionalized DNA nippers in a few minutes, which was applied as a 3D DNA nanomachine with an improved movement efficiency compared to traditional Au-based 3D nanomachines due to the organized and high local concentration of nippers on homogeneous DNA nanostructure. Once microRNA (miRNA) interacts with the 3D nanomachine, the nippers "open" to hybridize with the miRNA. Impressively, photoisomerization of the azo group induces dehybridization/hybridization of the nippers and miRNA under irradiation at different wavelengths, which easily solves one main technical challenge of DNA nanotechnology and biosensing: reversible locomotion in one step within 10 min. As a proof of concept, the described 3D machine is successfully applied in the rapid single-step detection of a biomarker, which gives impetus to the design of new generations of mechanical devices beyond the traditional ones with ultimate applications in sensing analysis and diagnostic technologies.
中文翻译:

高度有序且无场的 3D DNA 纳米结构:用于快速单步传感的下一代 DNA 纳米机器
在此,通过直接使用 Watson-Crick 碱基配对,偶氮苯 (azo) 功能化的 DNA 钳子在几分钟内自组装了高度有序且无场的三维 (3D) DNA 纳米结构,并将其作为 3D与传统的基于 Au 的 3D 纳米机器相比,DNA 纳米机器具有更高的运动效率,因为在均质 DNA 纳米结构上有组织且局部高度集中的钳子 一旦 microRNA (miRNA) 与 3D 纳米机器相互作用,钳子就会“打开”以与 miRNA 杂交。令人印象深刻的是,偶氮基团的光异构化在不同波长的照射下诱导了钳子和 miRNA 的脱杂交/杂交,这轻松解决了 DNA 纳米技术和生物传感的一项主要技术挑战:10 分钟内一步可逆运动。
更新日期:2018-07-16
中文翻译:

高度有序且无场的 3D DNA 纳米结构:用于快速单步传感的下一代 DNA 纳米机器
在此,通过直接使用 Watson-Crick 碱基配对,偶氮苯 (azo) 功能化的 DNA 钳子在几分钟内自组装了高度有序且无场的三维 (3D) DNA 纳米结构,并将其作为 3D与传统的基于 Au 的 3D 纳米机器相比,DNA 纳米机器具有更高的运动效率,因为在均质 DNA 纳米结构上有组织且局部高度集中的钳子 一旦 microRNA (miRNA) 与 3D 纳米机器相互作用,钳子就会“打开”以与 miRNA 杂交。令人印象深刻的是,偶氮基团的光异构化在不同波长的照射下诱导了钳子和 miRNA 的脱杂交/杂交,这轻松解决了 DNA 纳米技术和生物传感的一项主要技术挑战:10 分钟内一步可逆运动。











































 京公网安备 11010802027423号
京公网安备 11010802027423号