当前位置:
X-MOL 学术
›
Biochemistry
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
An Open Library of Human Kinase Domain Constructs for Automated Bacterial Expression
Biochemistry ( IF 2.9 ) Pub Date : 2018-07-13 00:00:00 , DOI: 10.1021/acs.biochem.7b01081 Steven K. Albanese 1, 2 , Daniel L. Parton 2 , Mehtap Işık 2, 3 , Lucelenie Rodríguez-Laureano 2 , Sonya M. Hanson 2 , Julie M. Behr 2, 4 , Scott Gradia 5 , Chris Jeans 5 , Nicholas M. Levinson 6 , Markus A. Seeliger 7 , John D. Chodera 2
Biochemistry ( IF 2.9 ) Pub Date : 2018-07-13 00:00:00 , DOI: 10.1021/acs.biochem.7b01081 Steven K. Albanese 1, 2 , Daniel L. Parton 2 , Mehtap Işık 2, 3 , Lucelenie Rodríguez-Laureano 2 , Sonya M. Hanson 2 , Julie M. Behr 2, 4 , Scott Gradia 5 , Chris Jeans 5 , Nicholas M. Levinson 6 , Markus A. Seeliger 7 , John D. Chodera 2
Affiliation
|
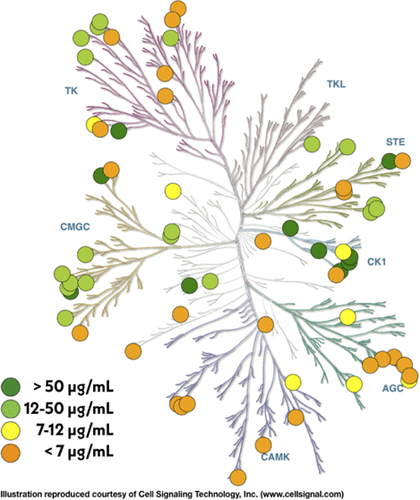
Kinases play a critical role in cellular signaling and are dysregulated in a number of diseases, such as cancer, diabetes, and neurodegeneration. Therapeutics targeting kinases currently account for roughly 50% of cancer drug discovery efforts. The ability to explore human kinase biochemistry and biophysics in the laboratory is essential to designing selective inhibitors and studying drug resistance. Bacterial expression systems are superior to insect or mammalian cells in terms of simplicity and cost effectiveness but have historically struggled with human kinase expression. Following the discovery that phosphatase coexpression produced high yields of Src and Abl kinase domains in bacteria, we have generated a library of 52 His-tagged human kinase domain constructs that express above 2 μg/mL of culture in an automated bacterial expression system utilizing phosphatase coexpression (YopH for Tyr kinases and lambda for Ser/Thr kinases). Here, we report a structural bioinformatics approach to identifying kinase domain constructs previously expressed in bacteria and likely to express well in our protocol, experiments demonstrating our simple construct selection strategy selects constructs with good expression yields in a test of 84 potential kinase domain boundaries for Abl, and yields from a high-throughput expression screen of 96 human kinase constructs. Using a fluorescence-based thermostability assay and a fluorescent ATP-competitive inhibitor, we show that the highest-expressing kinases are folded and have well-formed ATP binding sites. We also demonstrate that these constructs can enable characterization of clinical mutations by expressing a panel of 48 Src and 46 Abl mutations. The wild-type kinase construct library is available publicly via Addgene.
中文翻译:

一个开放的人激酶域结构的细菌自动表达库。
激酶在细胞信号传导中起关键作用,并且在许多疾病中失调,例如癌症,糖尿病和神经变性。目前,靶向激酶的治疗药物约占癌症药物发现努力的50%。在实验室中探索人激酶生物化学和生物物理学的能力对于设计选择性抑制剂和研究耐药性至关重要。就简单性和成本效益而言,细菌表达系统优于昆虫或哺乳动物细胞,但历史上一直在与人类激酶表达作斗争。在发现磷酸酶共表达可在细菌中产生高产率的Src和Abl激酶结构域之后,我们已经建立了一个52个带有His标签的人类激酶结构域构建体的文库,这些构建体在利用磷酸酶共表达的自动细菌表达系统中表达2μg/ mL以上的培养物(Tyr激酶为YopH,Ser / Thr激酶为lambda)。在这里,我们报告了一种结构生物信息学方法,用于鉴定以前在细菌中表达并可能在我们的方案中很好表达的激酶结构域构建体,实验证明了我们的简单构建体选择策略可以在对84个潜在的Abl激酶结构域边界进行测试的过程中选择具有良好表达产量的构建体,并从96种人激酶构建体的高通量表达筛选中获得收益。使用基于荧光的热稳定性测定法和具有荧光ATP竞争性的抑制剂,我们表明,表达最高的激酶被折叠并具有格式良好的ATP结合位点。我们还证明了这些构建体可以通过表达一组48 Src和46 Abl突变来表征临床突变。野生型激酶构建体文库可通过Addgene公开获得。
更新日期:2018-07-13
中文翻译:

一个开放的人激酶域结构的细菌自动表达库。
激酶在细胞信号传导中起关键作用,并且在许多疾病中失调,例如癌症,糖尿病和神经变性。目前,靶向激酶的治疗药物约占癌症药物发现努力的50%。在实验室中探索人激酶生物化学和生物物理学的能力对于设计选择性抑制剂和研究耐药性至关重要。就简单性和成本效益而言,细菌表达系统优于昆虫或哺乳动物细胞,但历史上一直在与人类激酶表达作斗争。在发现磷酸酶共表达可在细菌中产生高产率的Src和Abl激酶结构域之后,我们已经建立了一个52个带有His标签的人类激酶结构域构建体的文库,这些构建体在利用磷酸酶共表达的自动细菌表达系统中表达2μg/ mL以上的培养物(Tyr激酶为YopH,Ser / Thr激酶为lambda)。在这里,我们报告了一种结构生物信息学方法,用于鉴定以前在细菌中表达并可能在我们的方案中很好表达的激酶结构域构建体,实验证明了我们的简单构建体选择策略可以在对84个潜在的Abl激酶结构域边界进行测试的过程中选择具有良好表达产量的构建体,并从96种人激酶构建体的高通量表达筛选中获得收益。使用基于荧光的热稳定性测定法和具有荧光ATP竞争性的抑制剂,我们表明,表达最高的激酶被折叠并具有格式良好的ATP结合位点。我们还证明了这些构建体可以通过表达一组48 Src和46 Abl突变来表征临床突变。野生型激酶构建体文库可通过Addgene公开获得。



























 京公网安备 11010802027423号
京公网安备 11010802027423号